Many causes of autism, including environmental and genetic factors, have been recognized or proposed, but understanding of the theory of causation of autism is incomplete.[1] Attempts have been made to incorporate the known genetic and environmental causes into a comprehensive causative framework.[2] ASD (autism spectrum disorder) is a neurodevelopmental disorder marked by impairments in communicative ability and social interaction, as well as restricted and repetitive behaviors, interests, or activities not suitable for the individual's developmental stage. The severity of symptoms and functional impairment vary between individuals.[3]
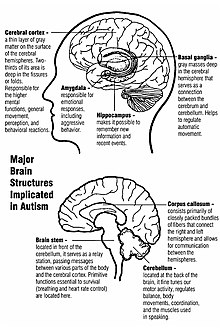
There are many known environmental, genetic, and biological causes of autism. Research indicates that genetic factors predominantly contribute to its appearance. The heritability of autism is complex and many of the genetic interactions involved are unknown.[1] In rare cases, autism has been associated with agents that cause birth defects.[4] Many other causes have been proposed.
Different underlying brain dysfunctions have been hypothesized to result in the common symptoms of autism, just as completely different brain types result in intellectual disability.[1][5] In recent years, the prevalence and number of people diagnosed with the disorder have increased dramatically. There are many potential reasons for this occurrence, particularly the changes in the diagnostic criteria for autism.[6]
Environmental factors that have been claimed to contribute to autism or exacerbate its symptoms, or that may be important to consider in future research, include certain foods,[7] infectious disease, heavy metals, solvents, diesel exhaust, PCBs, phthalates and phenols used in plastic products, pesticides, brominated flame retardants, alcohol, smoking, and illicit drugs.[6] Among these factors, vaccines have attracted much attention, as parents may first become aware of autistic symptoms in their child around the time of a routine vaccination, and parental concern about vaccines has led to a decreasing uptake of childhood immunizations and an increasing likelihood of measles outbreaks.[8][9] Overwhelming scientific evidence shows no causal association between the measles-mumps-rubella (MMR) vaccine and autism. Although there is no definitive evidence that the vaccine preservative thimerosal causes autism, studies have indicated a possible link between thimerosal and autism in individuals with a hereditary predisposition for autoimmune disorders.[10][11] In 2007, the Center for Disease Control stated there was no support for a link between thimerosal and autism, citing evidence from several studies, as well as a continued increase in autism cases following the removal of thimerosal from childhood vaccines.[12]
Genetics
editGenetic factors may be the most significant cause of autism. Early studies of twins had estimated heritability to be over 90%, meaning that genetics explains over 90% of whether a child will develop autism.[1] This may be an overestimation, as later twin studies estimate the heritability at between 60 and 90%.[1][13] Evidence so far still suggests a strong genetic component, with one of the largest and most recent studies estimating the heritability at 83%.[14] Many of the non-autistic co-twins had learning or social disabilities. For adult siblings the risk for having one or more features of the broader autism phenotype might be as high as 30%.[15]
In spite of the strong heritability, most cases of autism occur sporadically with no recent evidence of family history. It has been hypothesized that spontaneous de novo mutations in the sperm or egg contribute to the likelihood of developing autism.[16][1] There are two lines of evidence that support this hypothesis. First, individuals with autism have significantly reduced fecundity, they are 20 times less likely to have children than average, thus curtailing the persistence of mutations in ASD genes over multiple generations in a family.[1][5] Second, the likelihood of having a child develop autism increases with advancing parental age, and mutations in sperm gradually accumulate throughout a man's life.[1][17]
The first genes to be definitively shown to contribute to risk for autism were found in the early 1990s by researchers looking at gender-specific forms of autism caused by mutations on the X chromosome. An expansion of the CGG trinucleotide repeat in the promoter of the gene FMR1 in boys causes fragile X syndrome, and at least 20% of boys with this mutation have behaviors consistent with autism spectrum disorder.[18][19] Mutations that inactivate the gene MECP2 cause Rett syndrome, which is associated with autistic behaviors in girls, and in boys the mutation is embryonic lethal.[20]
Besides these early examples, the role of de novo mutations in autism first became evident when DNA microarray technologies reached sufficient resolution to allow the detection of copy number variation (CNV) in the human genome.[21][22] CNVs are the most common type of structural variation in the genome, consisting of deletions and duplications of DNA that range in size from a kilobase to a few megabases. Microarray analysis has shown that de novo CNVs occur at a significantly higher rate in sporadic cases of autism as compared to the rate in their typically developing siblings and unrelated controls. A series of studies have shown that gene disrupting de novo CNVs occur approximately four times more frequently in autism than in controls and contribute to approximately 5–10% of cases.[16][23][24][25] Based on these studies, there are predicted to be 130–234 autism-related CNV loci.[25] The first whole genome sequencing study to comprehensively catalog de novo structural variation at a much higher resolution than DNA microarray studies has shown that the mutation rate is approximately 20% and not elevated in autism compared to sibling controls.[26] Structural variants in individuals with autism are much larger and four times more likely to disrupt genes, mirroring findings from CNV studies.[26]
CNV studies were closely followed by exome sequencing studies, which sequence the 1–2% of the genome that codes for proteins (the "exome"). These studies found that de novo gene inactivating mutations were observed in approximately 20% of individuals with autism, compared to 10% of unaffected siblings, suggesting the etiology of autism is driven by these mutations in around 10% of cases.[27][28][29][30][31][32] There are predicted to be 350-450 genes that significantly increase susceptibility to autism when impacted by inactivating de novo mutations.[33] A further 12% of cases are predicted to be caused by protein altering missense mutations that change an amino acid but do not inactivate a gene.[29] Therefore, approximately 30% of individuals with autism have a spontaneous de novo large CNV that deletes or duplicates genes, or mutation that changes the amino acid code of an individual gene. A further 5–10% of cases have inherited structural variation at loci known to be associated with autism, and these known structural variants may arise de novo in the parents of affected children.[26]
Tens of genes and CNVs have been definitively identified based on the observation of recurrent mutations in different individuals, and suggestive evidence has been found for over 100 others.[34] The Simons Foundation Autism Research Initiative (SFARI) details the evidence for each genetic locus associated with autism.[35]
These early gene and CNV findings have shown that the cognitive and behavioral features associated with each of the underlying mutations is variable. Each mutation is itself associated with a variety of clinical diagnoses, and can also be found in a small percentage of individuals with no clinical diagnosis.[36][37] Thus the genetic disorders that comprise autism are not autism-specific. The mutations themselves are characterized by considerable variability in clinical outcome and typically only a subset of mutation carriers meet criteria for autism. This variable expressivity results in different individuals with the same mutation varying considerably in the severity of their observed particular trait.[38]
The conclusion of these recent studies of de novo mutation is that the spectrum of autism is breaking up into quanta of individual disorders defined by genetics.[38]
One gene that has been linked to autism is SHANK2.[39] Mutations in this gene act in a dominant fashion. Mutations in this gene appear to cause hyperconnectivity between the neurons.
A study conducted on 42,607 autism cases has identified 60 new genes, five of which had a more moderate impact on autistic symptoms. The related gene variants were often inherited from the participant's parents.[40]
Disorders
editSome conditions which may rarely be associated with an ASD appearance are:[41]
Amino acid metabolism
editY-aminobutyric acid metabolism
editCholesterol metabolism
editCerebral folate deficiency
edit- Folate receptor 1 gene mutations
- Dihydrofolate reductase deficiency
Creatine transport or metabolism
edit- Arginine:glycine amidinotransferase deficiency
- Guanidinoacetate methyltransferase deficiency
- X-linked creatine transporter defect
Carnitine biosynthesis
edit- 6-N-trimethyllysine dioxygenase deficiency
Purine and pyrimidine metabolism
edit- Adenylosuccinate lyase deficiency
- Adenosine deaminase deficiency
- Cytosolic 5'-nucleotidase superactivity
- Dihydropyrimidine dehydrogenase deficiency
- Phosphoribosyl pyrophosphate synthetase superactivity
Lysosomal storage
edit- Sanfilippo syndrome (mucopolysaccharidosis type III)
DNA
edit- Mitochondrial disease
- Nuclear DNA mutations
Biotinidase and urea
edit- Biotinidase deficiency
- Urea cycle defects
Epigenetics
editEpigenetic mechanisms may increase the risk of autism. Epigenetic changes occur as a result not of DNA sequence changes but of chromosomal histone modification or modification of the DNA bases. Such modifications are known to be affected by environmental factors, including nutrition, drugs, and mental stress.[42] Interest has been expressed in imprinted regions on chromosomes 15q and 7q.[43]
Most data supports a polygenic, epistatic model, meaning that the disorder is caused by two or more genes and that those genes are interacting in a complex manner. Several genes, between two and fifteen in number, have been identified and could potentially contribute to disease susceptibility.[44][45] An exact determination of the cause of ASD has yet to be discovered and there probably is not one single genetic cause of any particular set of disorders, leading many researchers to believe that epigenetic mechanisms, such as genomic imprinting or epimutations, may play a major role.[46][47]
Epigenetic mechanisms can contribute to disease phenotypes. Epigenetic modifications include DNA cytosine methylation and post-translational modifications to histones. These mechanisms contribute to regulating gene expression without changing the sequence of the DNA and may be influenced by exposure to environmental factors and may be heritable from parents.[43] Rett syndrome and Fragile X syndrome (FXS) are single gene disorders related to autism with overlapping symptoms that include deficient neurological development, impaired language and communication, difficulties in social interactions, and stereotyped hand gestures. It is not uncommon for a patient to be diagnosed with both autism and Rett syndrome and/or FXS. Epigenetic regulatory mechanisms play the central role in pathogenesis of these two disorders.[46][48][49]
Genomic imprinting may also contribute to the development of autism. Genomic imprinting is another example of epigenetic regulation of gene expression. In this instance, the epigenetic modification(s) causes the offspring to express the maternal copy of a gene or the paternal copy of a gene, but not both. The imprinted gene is silenced through epigenetic mechanisms. Candidate genes and susceptibility alleles for autism are identified using a combination of techniques, including genome-wide and targeted analyses of allele sharing in sib-pairs, using association studies and transmission disequilibrium testing (TDT) of functional and/or positional candidate genes and examination of novel and recurrent cytogenetic aberrations. Results from numerous studies have identified several genomic regions known to be subject to imprinting, candidate genes, and gene-environment interactions. Particularly, chromosomes 15q and 7q appear to be epigenetic hotspots in contributing to autism. Also, genes on the X chromosome may play an important role, as in Rett Syndrome.[43]
An important basis for autism causation is also the over- or underproduction of brain permanent cells (neurons, oligodendrocytes, and astrocytes) by the neural precursor cells during fetal development.[50]
Prenatal environment
editThe development of autism is associated with several prenatal risk factors, including advanced age in either parent, diabetes, bleeding, and maternal use of antibiotics and psychiatric drugs during pregnancy.[1][51][52] Autism has been linked to birth defect agents acting during the first eight weeks from conception, though these cases are rare.[53] If the mother of the child is dealing with autoimmune conditions or disorders while pregnant, it may have an effect on the child's development of autism.[54] All of these factors can cause inflammation or impair immune signaling in one way or another.[54]
Obstructive sleep apnea in pregnancy
editSleep apnea can result in intermittent hypoxia and has been increasing in prevalence due in part to the obesity epidemic. The known maternal risk factors for autism diagnosis in her offspring are similar to the risk factors for sleep apnea. For example, advanced maternal age, maternal obesity, maternal type 2 diabetes and maternal hypertension all increase the risk of autism in her offspring.[55][56][57][58] Likewise, these are all known risk factors for sleep apnea.[59][60][61]
One study found that gestational sleep apnea was associated with low reading test scores in children and that this effect may be mediated by an increased risk of the child having sleep apnea themselves.[62] Another study reported low social development scores in 64% of infants born to mothers with sleep apnea compared to 25% of infants born to controls, suggesting sleep apnea in pregnancy may have an effect on offspring neurodevelopment.[63] There was also an increase in the amount of snoring the mothers with sleep apnea reported in their infants when compared to controls.[63] Children with sleep apnea have "hyperactivity, attention problems, aggressivity, lower social competency, poorer communication, and/or diminished adaptive skills".[64] One study found significant improvements in ADHD-like symptoms, aggression, social problems and thought problems in autistic children who underwent adenotonsillectomy for sleep apnea.[65] Sleep problems in autism have been linked in a study to brain changes, particularly in the hippocampus, though this study does not prove causation.[66] A common presentation of sleep apnea in children with autism is insomnia.[67] All known genetic syndromes which are linked to autism have a high prevalence of sleep apnea. The prevalence of sleep apnea in Down's Syndrome is 50% - 100%.[68] Sleep problems and OSA in this population have been linked to language development.[69] Since autism manifests in the early developmental period, sleep apnea in Down's Syndrome and other genetic syndromes such as Fragile X start early (at infancy or shortly after), and sleep disturbances alter brain development,[70] it's plausible that some of the neurodevelopmental differences seen in these genetic syndromes are at least partially caused by the effects of untreated sleep apnea.
Infectious hypotheses
editOne hypothesis suggests that prenatal viral infection may contribute to the development of autism. Prenatal exposure to rubella or cytomegalovirus activates the mother's immune response and may greatly increase the risk for autism in mice.[71] Congenital rubella syndrome is the most convincing environmental cause of autism.[72] Infection-associated immunological events in early pregnancy may affect neural development more than infections in late pregnancy, not only for autism, but also for psychiatric disorders of presumed neurodevelopmental origin, notably schizophrenia.[73]
A 2021 meta-analysis of 36 studies suggested a relationship between mothers recalling an infection during pregnancy and having children with autism.[74]
Environmental agents
editTeratogens are environmental agents that cause birth defects. Some agents that are theorized to cause birth defects have also been suggested as potential autism risk factors, although there is little to no scientific evidence to back such claims. These include exposure of the embryo to valproic acid,[1] paracetamol,[75] thalidomide or misoprostol.[76] These cases are rare.[77] Questions have also been raised whether ethanol (grain alcohol) increases autism risk, as part of fetal alcohol syndrome or alcohol-related birth defects.[76] All known teratogens appear to act during the first eight weeks from conception, and though this does not exclude the possibility that autism can be initiated or affected later, it is strong evidence that autism arises very early in development.[4]
A small significant link was shown to exist between prenatal exposure to airborne pollutants and autism risk. This finding was not consistent across studies, and exposure to pollutants was measured indirectly.[78]
Autoimmune and inflammatory diseases
editMaternal inflammatory and autoimmune diseases can damage embryonic and fetal tissues, aggravating a genetic problem or damaging the nervous system.[79]
Other maternal conditions
editThyroid problems that lead to thyroxine deficiency in the mother in weeks 8–12 of pregnancy have been postulated to produce changes in the fetal brain leading to autism. Thyroxine deficiencies can be caused by inadequate iodine in the diet, and by environmental agents that interfere with iodine uptake or act against thyroid hormones. Possible environmental agents include flavonoids in food, tobacco smoke, and most herbicides. This hypothesis has not been tested.[80]
Diabetes in the mother during pregnancy is a significant risk factor for autism; a 2009 meta-analysis found that gestational diabetes was associated with a twofold increased risk. A 2014 review also found that maternal diabetes was significantly associated with an increased risk of autism.[55] Although diabetes causes metabolic and hormonal abnormalities and oxidative stress, no biological mechanism is known for the association between gestational diabetes and autism risk.[81]
Maternal diagnoses of polycystic ovary syndrome was found to associated with higher risk of autism.[82]
Maternal obesity during pregnancy may also increase the risk of autism, although further study is needed.[83]
Maternal malnutrition during preconception and pregnancy influences fetal neurodevelopment. Intrauterine growth restriction is associated with autism, in both term and preterm infants.[84]
Other in utero
editIt has been hypothesized that folic acid taken during pregnancy could play a role in reducing cases of autism by modulating gene expression through an epigenetic mechanism. This hypothesis is supported by multiple studies.[85]
Prenatal stress, consisting of exposure to life events or environmental factors that distress an expectant mother, has been hypothesized to contribute to autism, possibly as part of a gene-environment interaction. Autism has been reported to be associated with prenatal stress both with retrospective studies that examined stressors such as job loss and family discord, and with natural experiments involving prenatal exposure to storms; animal studies have reported that prenatal stress can disrupt brain development and produce behaviors resembling symptoms of autism.[86] Other studies cast doubt on this association, notably population based studies in England and Sweden finding no link between stressful life events and autism.[87]
The fetal testosterone theory hypothesizes that higher levels of testosterone in the amniotic fluid of mothers pushes brain development towards improved ability to see patterns and analyze complex systems while diminishing communication and empathy, emphasizing "male" traits over "female", or in E-S theory terminology, emphasizing "systemizing" over "empathizing". One project has published several reports suggesting that high levels of fetal testosterone could produce behaviors relevant to those seen in autism.[88]
Based in part on animal studies, diagnostic ultrasounds administered during pregnancy have been hypothesized to increase the child's risk of autism. This hypothesis is not supported by independently published research, and examination of children whose mothers received an ultrasound has failed to find evidence of harmful effects.[89]
Some research suggests that maternal exposure to selective serotonin reuptake inhibitors during pregnancy is associated with an increased risk of autism, but it remains unclear whether there is a causal link between the two.[90] There is evidence, for example, that this association may be an artifact of confounding by maternal mental illness.[91]
Paracetamol
editParacetamol (acetaminophen) use during pregnancy has been suggested as a possible risk factor for autism. A large prospective review of 2,480,797 children published in JAMA Pediatrics in April 2024 found "acetaminophen use during pregnancy was not associated with children’s risk of autism, ADHD, or intellectual disability in sibling control analysis".[92]
Perinatal environment
editAutism is associated with some perinatal and obstetric conditions. Infants that are born pre-term often have various neurodevelopmental impairments related to motor skills, cognition, receptive and expressive language, and socio-emotional capabilities.[93] Pre-term infants are also at a higher risk of having various neurodevelopmental disorders such as cerebral palsy and autism, as well as psychiatric disorders related to attention, anxiety, and impaired social communication.[93] It has also been proposed that the functions of the hypothalamic-pituitary-adrenal axis and brain connectivity in pre-term infants may be affected by NICU-related stress resulting in deficits in emotional regulation and socio-emotional capabilities.[93] A 2019 analysis of perinatal and neonatal risk factors found that autism was associated with abnormal fetal positioning, umbilical cord complications, low 5-minute Apgar score, low birth weight and gestation duration, fetal distress, meconium aspiration syndrome, trauma or injury during birth, maternal hemorrhaging, multiple birth, feeding disorders, neonatal anemia, birth defects/malformation, incompatibility with maternal blood type, and jaundice/hyperbilirubinemia. These associations do not denote a causal relationship for any individual factor.[94] There is growing evidence that perinatal exposure to air pollution may be a risk factor for autism, although this evidence has methodological limitations, including a small number of studies and failure to control for potential confounding factors.[95][96] A few studies have found an association between autism and frequent use of acetaminophen (e.g. Tylenol, Paracetamol) by the mother during pregnancy.[97][98] This association does not necessarily demonstrate a causal relationship.
Postnatal environment
editA wide variety of postnatal contributors to autism have been proposed, including gastrointestinal or immune system abnormalities, allergies, and exposure of children to drugs, infection, certain foods, or heavy metals. The evidence for these risk factors is anecdotal and has not been confirmed by reliable studies.[99]
Amygdala neurons
editThis theory hypothesizes that an early developmental failure involving the amygdala cascades on the development of cortical areas that mediate social perception in the visual domain. The fusiform face area of the ventral stream is implicated. The idea is that it is involved in social knowledge and social cognition, and that the deficits in this network are instrumental in causing autism.[100]
Autoimmune disease
editThis theory hypothesizes that autoantibodies that target the brain or elements of brain metabolism may cause or exacerbate autism. It is related to the maternal infection theory, except that it postulates that the effect is caused by the individual's own antibodies, possibly due to an environmental trigger after birth. It is also related to several other hypothesized causes; for example, viral infection has been hypothesized to cause autism via an autoimmune mechanism.[101]
Interactions between the immune system and the nervous system begin early during embryogenesis, and successful neurodevelopment depends on a balanced immune response. It is possible that aberrant immune activity during critical periods of neurodevelopment is part of the mechanism of some forms of autism.[102] A small percentage of autism cases are associated with infection, usually before birth. Results from immune studies have been contradictory. Some abnormalities have been found in specific subgroups, and some of these have been replicated. It is not known whether these abnormalities are relevant to the pathology of autism, for example, by infection or autoimmunity, or whether they are secondary to the disease processes.[103] As autoantibodies are found in diseases other than autism, and are not always present in autism,[104] the relationship between immune disturbances and autism remains unclear and controversial.[105] A 2015 systematic review and meta-analysis found that children with a family history of autoimmune diseases were at a greater risk of autism compared to children without such a history.[106]
When an underlying maternal autoimmune disease is present, antibodies circulating to the fetus could contribute to the development of autism spectrum disorders.[107]
Gastrointestinal connection
editGastrointestinal problems are one of the most commonly associated medical disorders in people with autism.[108] These are linked to greater social impairment, irritability, behavior and sleep problems, language impairments and mood changes, so the theory that they are an overlap syndrome has been postulated.[108][109] Studies indicate that gastrointestinal inflammation, food allergies, gluten-related disorders (celiac disease, wheat allergy, non-celiac gluten sensitivity), visceral hypersensitivity, dysautonomia and gastroesophageal reflux are the mechanisms that possibly link both.[109]
A 2016 review concludes that enteric nervous system abnormalities might play a role in several neurological disorders, including autism. Neural connections and the immune system are a pathway that may allow diseases originated in the intestine to spread to the brain.[110] A 2018 review suggests that the frequent association of gastrointestinal disorders and autism is due to abnormalities of the gut–brain axis.[108]
The "leaky gut syndrome" hypothesis developed by Andrew Wakefield, known for his fraudulent study on another cause of autism, is popular among parents of children with autism.[111][112][113] It is based on the idea that defects in the intestinal barrier produce an excessive increase in intestinal permeability, allowing substances present in the intestine (including bacteria, environmental toxins, and food antigens) to pass into the blood. The data supporting this theory are limited and contradictory, since both increased intestinal permeability and normal permeability have been documented in people with autism. Studies with mice provide some support to this theory and suggest the importance of intestinal flora, demonstrating that the normalization of the intestinal barrier was associated with an improvement in some of the autism-like behaviors.[110] Studies on subgroups of people with autism showed the presence of high plasma levels of zonulin, a protein that regulates permeability opening the "pores" of the intestinal wall, as well as intestinal dysbiosis (reduced levels of Bifidobacteria and increased abundance of Akkermansia muciniphila, Escherichia coli, Clostridia and Candida fungi that promote the production of proinflammatory cytokines, all of which produces excessive intestinal permeability.[114] This allows passage of bacterial endotoxins from the gut into the bloodstream, stimulating liver cells to secrete tumor necrosis factor alpha (TNFα), which modulates blood–brain barrier permeability. Studies on ASD people showed that TNFα cascades produce proinflammatory cytokines, leading to peripheral inflammation and activation of microglia in the brain, which indicates neuroinflammation.[114] In addition, neuroactive opioid peptides from digested foods have been shown to leak into the bloodstream and permeate the blood–brain barrier, influencing neural cells and causing autistic symptoms.[114] (See Endogenous opiate precursor theory)
After a preliminary 1998 study of three children with autism treated with secretin infusion reported improved GI function and dramatic improvement in behavior, many parents sought secretin treatment and a black market for the hormone developed quickly.[115] Later studies found secretin clearly ineffective in treating autism.[116]
Endogenous opiate precursor theory
editIn 1979, a possible association between autism and opiate was proposed, it was noted that injecting small amounts of opiates into young laboratory animals resulted in symptoms similar to those seen in autistic children.[117] The possibility of a relationship between autism and the consumption of gluten and casein was first articulated by Kalle Reichelt in 1991.[118]
Opiate theory hypothesizes that autism is the result of a metabolic disorder in which opioid peptides gliadorphin (aka gluteomorphin) and Casomorphin, produced through metabolism of gluten (present in wheat and related cereals) and casein (present in dairy products), pass through an abnormally permeable intestinal wall and then proceed to exert an effect on neurotransmission through binding with opioid receptors. It has been postulated that the resulting excess of opioids affects brain maturation and causes autistic symptoms including: behavioral difficulties, attention problems, and alterations in communicative capacity and social and cognitive functioning.[118][119]
Although high levels of these opioids are eliminated in the urine, it has been suggested that a small part of them cross into the brain causing interference of signal transmission and disruption of normal activity. Three studies have reported that urine samples of people with autism show an increased 24-hour peptide excretion.[118] A study with a control group found no appreciable differences in opioid levels in urine samples of people with autism compared to controls.[114] Two studies showed an increased opioid levels in cerebrospinal fluid of people with autism.[118]
The theory further states that removing opiate precursors from a child's diet may allow time for these behaviors to cease, and neurological development in very young children to resume normally.[120] As of 2021, reliable studies have not demonstrated the benefit of gluten-free diets in the treatment of autism.[121][7] In the subset of people who have gluten sensitivity there is limited evidence that suggests that a gluten-free diet may improve some autistic behaviors.[121][7]
Nutrition-related factors
editThere have been multiple attempts to uncover a link between various nutritional deficiencies such as vitamin D and folate and autism risk.[122] Although there have been many studies on the role of vitamin D in the development of autism, the majority of them are limited by their inability to assess the deficiency prior to an autism diagnosis.[122] A meta-analysis on the association between vitamin D and autism found that individuals with autism had significantly low levels of serum 25-hydroxy vitamin D than those without autism.[122] Another analysis showed significant differences in levels of zinc between individuals with and without autism. Although studies showed significant differences protein intake and calcium in individuals with autism, the results were limited by their imprecision, inconsistency, and indirect nature.[122] Additionally, low levels of 5-methyltetrahydrofolate (5-MTHF) in the brain can result in cerebral folate deficiency (CFD) which has been shown to be associated with autism.[122][123]
Toxic exposure
editMultiple studies have attempted to study the relationship between toxic exposure and autism, despite limitations related to the measurement of toxic exposure the methods for which were often indirect and cross-sectional. Systematic reviews have been conducted for numerous toxins including air pollution, thimerosal, inorganic mercury, and levels of heavy metals in hair, nails, and bodily fluids.[122]
Although no link was found to exist between the vaccine additive thiomersal and autism risk, this association may hold true for individuals with a hereditary predisposition for autoimmune disorders.[11][122]
Environmental exposure to inorganic mercury may be associated with higher autism risk, with high levels of mercury in the body being a valid disease-causing agent for autism.[122][124]
Significant evidence has not been found of an association between autism and the concentration of copper, cadmium, selenium, and chromium in the hair, nails, and bodily fluids.[122][125][124] Levels of lead were found to be significantly higher in individuals with autism.[122][124] The precision and consistency of results were not maintained across studies and were influenced by an outlier study.[122] The atypical eating behaviors of autistic children, along with habitual mouthing and pica, make it hard to determine whether increased lead levels are a cause or a consequence of autism.[126]
Locus coeruleus–noradrenergic system
editThis theory hypothesizes that autistic behaviors depend at least in part on a developmental dysregulation that results in impaired function of the locus coeruleus–noradrenergic (LC-NA) system. The LC-NA system is heavily involved in arousal and attention; for example, it is related to the brain's acquisition and use of environmental cues.[127]
Oxidative stress
editOxidative stress, oxidative DNA damage and disruptions of DNA repair have been postulated to play a role in the etiopathology of both ASD and schizophrenia.[128] Physiological factors and mechanisms influence by oxidative stress are believed to be highly influential to autism risk. Interactions between environmental and genetic factors may increase oxidative stress in children with autism.[129] This theory hypothesizes that toxicity and oxidative stress may cause autism in some cases. Evidence includes genetic effects on metabolic pathways, reduced antioxidant capacity, enzyme changes, and enhanced biomarkers for oxidative stress.[129] One theory is that stress damages Purkinje cells in the cerebellum after birth, and it is possible that glutathione is involved.[130] Polymorphism of genes involved metabolization of glutathione is evidenced by lower levels of total glutathione, and higher levels of oxidized glutathione in autistic children.[129][131] Based on this theory, antioxidants may be a useful treatment for autism.[132] Environmental factors can influence oxidative stress pre, peri, and postnatally and include heavy metals, infection, certain drugs, and toxic exposure from various sources including cigarette smoke, air pollutants, and organophosphate pesticides.[129]
Social construct
editBeyond the genetic, epigenetic, and biological factors that can contribute to an autism diagnosis are theories related to the "autistic identity".[133] It has been theorized that perceptions towards the characteristics of autistic individuals have been heavily influenced by neurotypical ideologies and social norms.[133]
The social construct theory says that the boundary between normal and abnormal is subjective and arbitrary, so autism does not exist as an objective entity, but only as a social construct. It further argues that autistic individuals themselves have a way of being that is partly socially constructed.[134]
Mild and moderate variations of autism are particular targets of the theory that social factors determine what it means to be autistic. The theory hypothesizes that individuals with these diagnoses inhabit the identities that have been ascribed to them, and promote their sense of well-being by resisting or appropriating autistic ascriptions.[135]
Lynn Waterhouse suggests that autism has been reified, in that social processes have endowed it with more reality than is justified by the scientific evidence.[136]
Although social construction of the autistic identity can have a positive impact on the well-being and treatment of autistic individuals.[133] That is not always the case when the individuals in question belong to historically marginalized populations.[133]
Viral infection
editMany studies have presented evidence for and against association of autism with viral infection after birth. Laboratory rats infected with Borna disease virus show some symptoms similar to those of autism but blood studies of autistic children show no evidence of infection by this virus. Members of the herpes virus family may have a role in autism, but the evidence so far is anecdotal. Viruses have long been suspected as triggers for immune-mediated diseases such as multiple sclerosis but showing a direct role for viral causation is difficult in those diseases, and mechanisms, whereby viral infections could lead to autism, are speculative.[71]
Evolutionary explanations
editResearch exploring the evolutionary benefits of autism and associated genes suggests that people with autistic traits may have made facilitated crucial advancements in technology and knowledge of natural systems in the course of human development.[137][138] It has been suggested that these trait advantages may have resulted from the exchange of socially beneficial traits with ones that promote technological skills and systematic thought processes. In future studies, autism may prove similar to diseases such as sickle cell anemia, that demonstrate balanced polymorphism.[139]
A 2011 study proposed the "Solitary Forager Hypothesis" in which autistic traits, including increased abilities for spatial intelligence, concentration and memory, could have been naturally selected to enable self-sufficient foraging in a more solitary environment.[140][141][142] The author notes that such individuals likely foraged by themselves while occasionally interacting with intimate people or groups. A study conducted by Spikins et al. (2016) examined the role of Asperger syndrome as "an alternative pro-social adaptive strategy", which may have developed as a result of the emergence of "collaborative morality" in the context of small-scale hunter-gathering. The authors further suggest that "mutual interdependence of different social strategies" may have "contributed to the rise of innovation and large scale social networks".[143]
Conversely, noting the failure to find specific alleles that reliably cause autism or rare mutations that account for more than 5% of the heritable variation in autism established by twin and adoption studies, research in evolutionary psychiatry has concluded that it is unlikely that there is or has been selection pressure for autism when considering that, like schizophrenics, autistic people and their siblings tend to have fewer offspring on average than non-autistic people, and instead that autism is probably better explained as a by-product of adaptive traits caused by antagonistic pleiotropy and by genes that are retained due to a fitness landscape with an asymmetric distribution.[144][145][146]
Neanderthal theory
editOne theory on the evolutionary and biological origins of autism traits in Homo sapiens that has gained recent attention in the 2010s and 2020s is that some genes linked to autism may have originated from early humans crossbreeding with Neanderthals, an extinct group of archaic humans (generally regarded as a distinct species, Homo neanderthalensis, though some regard it as a subspecies of Homo sapiens, referred to as H. sapiens neanderthalensis) who lived in Eurasia until about 40,000 years ago.
A possible link between autism spectrum disorders (ASDs) and Neanderthal DNA was identified in 2009, pending genome sequencing.[148]
The first Neanderthal genome sequence was published in 2010, and strongly indicated interbreeding between Neanderthals and early modern humans.[149][150][151][152] The genomes of all studied modern populations contain Neanderthal DNA.[149][153][154][155][156] Various estimates exist for the proportion, such as 1–4%[149] or 3.4–7.9% in modern Eurasians,[157] or 1.8–2.4% in modern Europeans and 2.3–2.6% in modern East Asians.[158] Pre-agricultural Europeans appear to have had similar, or slightly higher,[156] percentages to modern East Asians, and the numbers may have decreased in the former due to dilution with a group of people which had split off before Neanderthal introgression.[159]
Typically, studies have reported finding no significant levels of Neanderthal DNA in Sub-Saharan Africans, but a 2020 study detected 0.3-0.5% in the genomes of five African sample populations, likely the result of Eurasians back-migrating and interbreeding with Africans, as well as human-to-Neanderthal gene flow from dispersals of Homo sapiens preceding the larger Out-of-Africa migration, and also showed more equal Neanderthal DNA percentages for European and Asian populations.[156] Such low percentages of Neanderthal DNA in all present day populations indicate infrequent past interbreeding,[160] unless interbreeding was more common with a different population of modern humans which did not contribute to the present day gene pool.[159] Of the inherited Neanderthal genome, 25% in modern Europeans and 32% in modern East Asians may be related to viral immunity.[161] In all, approximately 20% of the Neanderthal genome appears to have survived in the modern human gene pool.[162]
Due to their small population and resulting reduced effectivity of natural selection, Neanderthals accumulated several weakly harmful mutations, which were introduced to and slowly selected out of the much larger modern human population; the initial hybridised population may have experienced up to a 94% reduction in fitness compared to contemporary humans. By this measure, Neanderthals may have substantially increased in fitness.[163] A 2017 study focusing on archaic genes in Turkey found associations with coeliac disease, malaria severity and Costello syndrome.[164]
Nonetheless, some genes may have helped modern East Asians adapt to the environment; the putatively Neanderthal Val92Met variant of the MC1R gene, which may be weakly associated with red hair and UV radiation sensitivity,[165] is primarily found in East Asian, rather than European, individuals.[166] Some genes related to the immune system appear to have been affected by introgression, which may have aided migration,[167] such as OAS1,[168] STAT2,[169] TLR6, TLR1, TLR10,[170] and several related to immune response.[171][a] In addition, Neanderthal genes have also been implicated in the structure and function of the brain,[b] keratin filaments, sugar metabolism, muscle contraction, body fat distribution, enamel thickness and oocyte meiosis.[173] Nonetheless, a large portion of surviving introgression appears to be non-coding ("junk") DNA with few biological functions.[159]
A 2016 study indicated that human-Neanderthal gene variance may be involved in autism, with chromosome 16 section 16p11.2 deletions playing a large role.[174][175]
A 2017 study reported finding that the more Neanderthal DNA a person has in their genome, the more closely the brain of the individual would resemble that of a Neanderthal. The study also found that parts of the Neanderthal brain related to tool use and visual discrimination may have also experienced evolutionary or adaptational "trade-offs" with the "social brain", as also found in scientific studies on autism.[176] A 2023 study also found evidence that Neanderthal single nucleotide polymorphisms (SNPs) likely play a "significant role" in autism susceptibility and heritability in autism populations across the United States. According to the study, "Although most studies on autism genomics focus on the deleterious nature of variants, there is the possibility some of these autism-associated Neanderthal SNPs have been under weak positive selection. In support, recent studies have identified genetic variants implicated in both autism and high intelligence. Meanwhile, autistic people often perform better on tests of fluid intelligence than neurotypicals."[citation needed]
Another 2017 study that analyzed 68 genes associated with neurodevelopmental disorders, including autism, found that these disorders were also affected by natural selection and interbreeding between Homo sapiens and other archaic human species. The study also recommended further research into the link between Neanderthal single nucleotide polymorphisms (SNPs) and neurodevelopmental disorders, including autism, in modern-day humans.[177]
A 2021 study confirmed these findings, noting that "the protective allele of rs7170637(A) CYFIP1, [one of the genes associated with autism spectrum disorder (ASD)], was present in primates to Neanderthals, and reemerged in modern humans, while absent in early modern humans"; "identified significant positive selection signals in 18 ASD risk SNPs"; that "ancient genome analysis identified de novo mutations...representing genes involved in cognitive function...and conserved evolutionary selection clusters"; and that "relative enrichment of the ASD risk SNPs from the respective evolutionary cluster or biological interaction networks may help in addressing the phenotypic diversity in ASD", with "cognitive genomic tradeoff signatures impacting the biological networks [explaining] the paradoxical phenotypes in ASD".[178]
Discredited theories
editRefrigerator mother
editPsychologist Bruno Bettelheim believed that autism was linked to early childhood trauma, and his work was highly influential for decades both in the medical and popular spheres. In his discredited theory, he blamed the mothers of individuals with autism for having caused their child's condition through the withholding of affection.[179] Leo Kanner, who first described autism,[180] suggested that parental coldness might contribute to autism.[181] Although Kanner eventually renounced the theory, Bettelheim put an almost exclusive emphasis on it in both his medical and his popular books. Treatments based on these theories failed to help children with autism, and after Bettelheim's death, his reported rates of cure (around 85%) were found to be fraudulent.[182]
Vaccines
editThe most recent scientific research has determined that changes to brain structures correlated with the development of autism can already be detected while the child is still in the womb, well before any vaccines are administered.[183] Furthermore, scientific studies have consistently refuted a causal relationship between vaccinations and autism.[184][185][186]
Despite this, some parents believe that vaccinations cause autism; they therefore delay or avoid immunizing their children (for example, under the "vaccine overload" hypothesis that giving many vaccines at once may overwhelm a child's immune system and lead to autism,[187] even though this hypothesis has no scientific evidence and is biologically implausible[188]). Diseases such as measles can cause severe disabilities and even death, so the risk of death or disability for an unvaccinated child is higher than the risk for a child who has been vaccinated.[189] Despite medical evidence, antivaccine activism continues. A developing tactic is the "promotion of irrelevant research to justify the science underlying a questionable claim."[190]
MMR vaccine
editThe MMR vaccine as a cause of autism is one of the most extensively debated hypotheses regarding the origins of autism. Andrew Wakefield et al. reported a study of 12 children who had autism and bowel symptoms, in some cases reportedly with onset after MMR.[191] Although the paper, which was later retracted by the journal, concluded that there was no association between the MMR vaccine and autism, Wakefield nevertheless suggested a false notion during a 1998 press conference that giving children the vaccines in three separate doses would be safer than a single dose.[191][192] Administering the vaccines in three separate doses does not reduce the chance of adverse effects, and it increases the opportunity for infection by the two diseases not immunized against first.[8][10]
In 2004, the interpretation of a causal link between MMR vaccine and autism was formally retracted by ten of Wakefield's twelve co-authors.[193] The retraction followed an investigation by The Sunday Times, which stated that Wakefield "acted dishonestly and irresponsibly".[194] The Centers for Disease Control and Prevention, the Institute of Medicine of the National Academy of Sciences, and the U.K. National Health Service have all concluded that there is no evidence of a link between the MMR vaccine and autism.[195][196][197]
In February 2010, The Lancet, which published Wakefield's study, fully retracted it after an independent auditor found the study to be flawed.[191] In January 2011, an investigation published in the journal BMJ described the Wakefield study as the result of deliberate fraud and manipulation of data.[198][199][200][201]
Thiomersal (thimerosal)
editPerhaps the best-known hypothesis involving mercury and autism involves the use of the mercury-based compound thiomersal, a preservative that has been phased out from most childhood vaccinations in developed countries including US and the EU.[202] There is no scientific evidence for a causal connection between thiomersal and autism, but parental concern about a relationship between thiomersal and vaccines has led to decreasing rates of childhood immunizations and increasing likelihood of disease outbreaks.[8][9][10] In 1999, due to concern about the dose of mercury infants were being exposed to, the U.S. Public Health Service recommended that thiomersal be removed from childhood vaccines, and by 2002 the flu vaccine was the only childhood vaccine containing more than trace amounts of thimerosal. Despite this, autism rates did not decrease after the removal of thimerosal, in the US or other countries that also removed thimerosal from their childhood vaccines.[203]
A causal link between thimerosal and autism has been rejected by international scientific and medical professional bodies including the American Medical Association,[204] the American Academy of Pediatrics,[205] the American College of Medical Toxicology,[206] the Canadian Paediatric Society,[207] the U.S. National Academy of Sciences,[196] the Food and Drug Administration,[208] Centers for Disease Control and Prevention,[195] the World Health Organization,[209] the Public Health Agency of Canada,[210] and the European Medicines Agency.[211]
See also
editNotes
edit- ^ OAS1[168] and STAT2[169] both are associated with fighting viral inflections (interferons), and the listed toll-like receptors (TLRs)[170] allow cells to identify bacterial, fungal, or parasitic pathogens. African origin is also correlated with a stronger inflammatory response.[171]
- ^ Higher levels of Neanderthal-derived genes are associated with an occipital and parietal bone shape reminiscent to that of Neanderthals, as well as modifications to the visual cortex and the intraparietal sulcus (associated with visual processing).[172]
References
edit- ^ a b c d e f g h i j Waye MM, Cheng HY (April 2018). "Genetics and epigenetics of autism: A Review". Psychiatry and Clinical Neurosciences (Review). 72 (4): 228–244. doi:10.1111/pcn.12606. eISSN 1440-1819. PMID 28941239. S2CID 206257210.
- ^ Sarovic D (November 2021). "A Unifying Theory for Autism: The Pathogenetic Triad as a Theoretical Framework". Frontiers in Psychiatry (Review). 12: 767075. doi:10.3389/fpsyt.2021.767075. PMC 8637925. PMID 34867553. S2CID 244119594.
- ^ American Psychiatric Association (2022). Diagnostic and statistical manual of mental disorders: DSM-5-TR (5th ed.). American Psychiatric Association Publishing. doi:10.1176/appi.books.9780890425787. ISBN 978-0-89042-575-6. S2CID 249488050.
- ^ a b Arndt TL, Stodgell CJ, Rodier PM (2005). "The teratology of autism". International Journal of Developmental Neuroscience (Review). 23 (2–3): 189–199. doi:10.1016/j.ijdevneu.2004.11.001. PMID 15749245. S2CID 17797266.
- ^ a b Hodges H, Fealko C, Soares N (February 2020). "Autism spectrum disorder: definition, epidemiology, causes, and clinical evaluation". Translational Pediatrics. 9 (Suppl 1): S55–S65. doi:10.21037/tp.2019.09.09. PMC 7082249. PMID 32206584.
- ^ a b Salari N, Rasoulpoor S, Rasoulpoor S, Shohaimi S, Jafarpour S, Abdoli N, et al. (July 2022). "The global prevalence of autism spectrum disorder: a comprehensive systematic review and meta-analysis". Italian Journal of Pediatrics. 48 (1): 112. doi:10.1186/s13052-022-01310-w. PMC 9270782. PMID 35804408.
- ^ a b c Quan J, Panaccione N, King JA, Underwood F, Windsor JW, Coward S, et al. (March 2019). "A257 Association Between Celiac Disease and Autism Spectrum Disorder: A Systematic Review". Journal of the Canadian Association of Gastroenterology. 2 (Supplement_2): 502–503. doi:10.1093/jcag/gwz006.256. ISSN 2515-2084. PMC 6512700.
- ^ a b c Wilder-Smith AB, Qureshi K (March 2020). "Resurgence of Measles in Europe: A Systematic Review on Parental Attitudes and Beliefs of Measles Vaccine". Journal of Epidemiology and Global Health. 10 (1): 46–58. doi:10.2991/jegh.k.191117.001. PMC 7310814. PMID 32175710.
- ^ a b Gidengil C, Chen C, Parker AM, Nowak S, Matthews L (October 2019). "Beliefs around childhood vaccines in the United States: A systematic review". Vaccine. 37 (45): 6793–6802. doi:10.1016/j.vaccine.2019.08.068. PMC 6949013. PMID 31562000.
- ^ a b c Di Pietrantonj C, Rivetti A, Marchione P, Debalini MG, Demicheli V, et al. (Cochrane Acute Respiratory Infections Group) (November 2021). "Vaccines for measles, mumps, rubella, and varicella in children". The Cochrane Database of Systematic Reviews. 2021 (11): CD004407. doi:10.1002/14651858.CD004407.pub5. PMC 8607336. PMID 34806766.
- ^ a b Kern JK, Geier DA, Mehta JA, Homme KG, Geier MR (December 2020). "Mercury as a hapten: A review of the role of toxicant-induced brain autoantibodies in autism and possible treatment considerations". Journal of Trace Elements in Medicine and Biology. 62: 126504. Bibcode:2020JTEMB..6226504K. doi:10.1016/j.jtemb.2020.126504. PMID 32534375. S2CID 219468115.
- ^ "Timeline: Thimerosal in Vaccines (1999-2010)". CDC. 19 August 2020. Retrieved 2024-04-24.
- ^ Carlsson T, Molander F, Taylor MJ, Jonsson U, Bölte S (October 2021). "Early environmental risk factors for neurodevelopmental disorders - a systematic review of twin and sibling studies". Development and Psychopathology. 33 (4): 1448–1495. doi:10.1017/S0954579420000620. PMC 8564717. PMID 32703331.
- ^ Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A (September 2017). "The Heritability of Autism Spectrum Disorder". JAMA. 318 (12): 1182–1184. doi:10.1001/jama.2017.12141. PMC 5818813. PMID 28973605.
- ^ Folstein SE, Rosen-Sheidley B (December 2001). "Genetics of autism: complex aetiology for a heterogeneous disorder". Nature Reviews. Genetics (Review). 2 (12): 943–955. doi:10.1038/35103559. PMID 11733747. S2CID 9331084.
- ^ a b Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. (April 2007). "Strong association of de novo copy number mutations with autism". Science. 316 (5823): 445–449. Bibcode:2007Sci...316..445S. doi:10.1126/science.1138659. PMC 2993504. PMID 17363630.
- ^ Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, et al. (August 2012). "Rate of de novo mutations and the importance of father's age to disease risk". Nature. 488 (7412): 471–475. Bibcode:2012Natur.488..471K. doi:10.1038/nature11396. PMC 3548427. PMID 22914163.
- ^ Man L, Lekovich J, Rosenwaks Z, Gerhardt J (2017-09-12). "Fragile X-Associated Diminished Ovarian Reserve and Primary Ovarian Insufficiency from Molecular Mechanisms to Clinical Manifestations". Frontiers in Molecular Neuroscience. 10: 290. doi:10.3389/fnmol.2017.00290. PMC 5600956. PMID 28955201.
- ^ Hatton DD, Sideris J, Skinner M, Mankowski J, Bailey DB, Roberts J, Mirrett P (September 2006). "Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP". American Journal of Medical Genetics. Part A. 140A (17): 1804–1813. doi:10.1002/ajmg.a.31286. PMID 16700053. S2CID 11017841.
- ^ Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (October 1999). "Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2". Nature Genetics. 23 (2): 185–188. doi:10.1038/13810. PMID 10508514. S2CID 3350350.
- ^ Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. (July 2004). "Large-scale copy number polymorphism in the human genome". Science. 305 (5683): 525–528. Bibcode:2004Sci...305..525S. doi:10.1126/science.1098918. PMID 15273396. S2CID 20357402.
- ^ Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. (September 2004). "Detection of large-scale variation in the human genome". Nature Genetics. 36 (9): 949–951. doi:10.1038/ng1416. PMID 15286789. S2CID 1433674.
- ^ Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, et al. (May 2014). "Convergence of genes and cellular pathways dysregulated in autism spectrum disorders". American Journal of Human Genetics. 94 (5): 677–694. doi:10.1016/j.ajhg.2014.03.018. PMC 4067558. PMID 24768552.
- ^ Levy D, Ronemus M, Yamrom B, Lee YH, Leotta A, Kendall J, et al. (June 2011). "Rare de novo and transmitted copy-number variation in autistic spectrum disorders". Neuron. 70 (5): 886–897. doi:10.1016/j.neuron.2011.05.015. PMID 21658582. S2CID 11132936.
- ^ a b Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. (June 2011). "Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism". Neuron. 70 (5): 863–885. doi:10.1016/j.neuron.2011.05.002. PMC 3939065. PMID 21658581.
- ^ a b c Brandler WM, Antaki D, Gujral M, Noor A, Rosanio G, Chapman TR, et al. (April 2016). "Frequency and Complexity of De Novo Structural Mutation in Autism". American Journal of Human Genetics. 98 (4): 667–679. doi:10.1016/j.ajhg.2016.02.018. PMC 4833290. PMID 27018473.
- ^ Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. (April 2012). "De novo gene disruptions in children on the autistic spectrum". Neuron. 74 (2): 285–299. doi:10.1016/j.neuron.2012.04.009. PMC 3619976. PMID 22542183.
- ^ De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. (November 2014). "Synaptic, transcriptional and chromatin genes disrupted in autism". Nature. 515 (7526): 209–215. Bibcode:2014Natur.515..209.. doi:10.1038/nature13772. PMC 4402723. PMID 25363760.
- ^ a b Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. (November 2014). "The contribution of de novo coding mutations to autism spectrum disorder". Nature. 515 (7526): 216–221. Bibcode:2014Natur.515..216I. doi:10.1038/nature13908. PMC 4313871. PMID 25363768.
- ^ Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, et al. (April 2012). "Patterns and rates of exonic de novo mutations in autism spectrum disorders". Nature. 485 (7397): 242–245. Bibcode:2012Natur.485..242N. doi:10.1038/nature11011. PMC 3613847. PMID 22495311.
- ^ Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. (April 2012). "De novo mutations revealed by whole-exome sequencing are strongly associated with autism". Nature. 485 (7397): 237–241. Bibcode:2012Natur.485..237S. doi:10.1038/nature10945. PMC 3667984. PMID 22495306.
- ^ O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. (April 2012). "Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations". Nature. 485 (7397): 246–250. Bibcode:2012Natur.485..246O. doi:10.1038/nature10989. PMC 3350576. PMID 22495309.
- ^ Ronemus M, Iossifov I, Levy D, Wigler M (February 2014). "The role of de novo mutations in the genetics of autism spectrum disorders". Nature Reviews. Genetics. 15 (2): 133–141. doi:10.1038/nrg3585. PMID 24430941. S2CID 9073763.
- ^ Betancur C (March 2011). "Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting" (PDF). Brain Research. 1380: 42–77. doi:10.1016/j.brainres.2010.11.078. PMID 21129364. S2CID 41429306.
- ^ "SFARI Gene". SFARI gene. Archived from the original on 2016-04-01. Retrieved 2016-04-13.
- ^ Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, et al. (January 2014). "CNVs conferring risk of autism or schizophrenia affect cognition in controls". Nature. 505 (7483): 361–366. Bibcode:2014Natur.505..361S. doi:10.1038/nature12818. hdl:2336/311615. PMID 24352232. S2CID 3842341.
- ^ Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, et al. (May 2010). "Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size". Journal of Medical Genetics. 47 (5): 332–341. doi:10.1136/jmg.2009.073015. PMC 3158566. PMID 19914906.
- ^ a b Brandler WM, Sebat J (14 January 2015). "From de novo mutations to personalized therapeutic interventions in autism". Annual Review of Medicine. 66 (1): 487–507. doi:10.1146/annurev-med-091113-024550. PMID 25587659.
- ^ Zaslavsky K, Zhang WB, McCready FP, Rodrigues DC, Deneault E, Loo C, et al. (April 2019). "SHANK2 mutations associated with autism spectrum disorder cause hyperconnectivity of human neurons". Nature Neuroscience. 22 (4): 556–564. doi:10.1038/s41593-019-0365-8. PMC 6475597. PMID 30911184.
- ^ Columbia University Irving Medical Center (September 4, 2022). "60 New Genes Linked to Autism Uncovered". Nature Genetics. 54 (9). SciTech Daily: 1305–1319. doi:10.1038/s41588-022-01148-2. PMC 9470534. PMID 35982159. Retrieved September 7, 2022.
- ^ Hyman SL, Levy SE, Myers SM (January 2020). "Identification, Evaluation, and Management of Children With Autism Spectrum Disorder". Pediatrics. 145 (1): e20193447. doi:10.1542/peds.2019-3447. PMID 31843864. S2CID 209390456.
- ^ Miyake K, Hirasawa T, Koide T, Kubota T (2012). "Epigenetics in Autism and Other Neurodevelopmental Diseases". Neurodegenerative Diseases (Review). Advances in Experimental Medicine and Biology. Vol. 724. pp. 91–98. doi:10.1007/978-1-4614-0653-2_7. ISBN 978-1-4614-0652-5. PMID 22411236.
- ^ a b c Schanen NC (October 2006). "Epigenetics of autism spectrum disorders". Human Molecular Genetics (Review). 15 Spec No 2: R138–R150. doi:10.1093/hmg/ddl213. PMID 16987877.
- ^ Pickles A, Bolton P, Macdonald H, Bailey A, Le Couteur A, Sim CH, Rutter M (September 1995). "Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism". American Journal of Human Genetics. 57 (3): 717–726. PMC 1801262. PMID 7668301.
- ^ Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, et al. (August 1999). "A genomic screen of autism: evidence for a multilocus etiology". American Journal of Human Genetics. 65 (2): 493–507. doi:10.1086/302497. PMC 1377948. PMID 10417292.
- ^ a b Samaco RC, Hogart A, LaSalle JM (February 2005). "Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3". Human Molecular Genetics. 14 (4): 483–492. doi:10.1093/hmg/ddi045. PMC 1224722. PMID 15615769.
- ^ Jiang YH, Sahoo T, Michaelis RC, Bercovich D, Bressler J, Kashork CD, et al. (November 2004). "A mixed epigenetic/genetic model for oligogenic inheritance of autism with a limited role for UBE3A". American Journal of Medical Genetics. Part A. 131 (1): 1–10. doi:10.1002/ajmg.a.30297. PMID 15389703. S2CID 9570482.
- ^ Lopez-Rangel E, Lewis ME (2006). "Further evidence for pigenetic influence of MECP2 in Rett, autism and Angelman's syndromes". Clinical Genetics. 69 (1): 23–25. doi:10.1111/j.1399-0004.2006.00543c.x. S2CID 85160435.
- ^ Hagerman RJ, Ono MY, Hagerman PJ (September 2005). "Recent advances in fragile X: a model for autism and neurodegeneration". Current Opinion in Psychiatry. 18 (5): 490–496. doi:10.1097/01.yco.0000179485.39520.b0. PMID 16639106. S2CID 33650811.
- ^ Rutgers University (August 21, 2022). "Scientists Discover That Irregular Production of Brain Cells Could Cause Autism". SciTech Daily.
- ^ Lee E, Cho J, Kim KY (October 2019). "The Association between Autism Spectrum Disorder and Pre- and Postnatal Antibiotic Exposure in Childhood-A Systematic Review with Meta-Analysis". International Journal of Environmental Research and Public Health. 16 (20): 4042. doi:10.3390/ijerph16204042. PMC 6843945. PMID 31652518.
- ^ Morales DR, Slattery J, Evans S, Kurz X (January 2018). "Antidepressant use during pregnancy and risk of autism spectrum disorder and attention deficit hyperactivity disorder: systematic review of observational studies and methodological considerations". BMC Medicine. 16 (1): 6. doi:10.1186/s12916-017-0993-3. PMC 5767968. PMID 29332605.
- ^ Roullet FI, Lai JK, Foster JA (2013). "In utero exposure to valproic acid and autism--a current review of clinical and animal studies". Neurotoxicology and Teratology (Review). 36: 47–56. Bibcode:2013NTxT...36...47R. doi:10.1016/j.ntt.2013.01.004. PMID 23395807.
- ^ a b Han VX, Patel S, Jones HF, Nielsen TC, Mohammad SS, Hofer MJ, et al. (January 2021). "Maternal acute and chronic inflammation in pregnancy is associated with common neurodevelopmental disorders: a systematic review". Translational Psychiatry. 11 (1): 71. doi:10.1038/s41398-021-01198-w. PMC 7820474. PMID 33479207.
- ^ a b Xu G, Jing J, Bowers K, Liu B, Bao W (April 2014). "Maternal diabetes and the risk of autism spectrum disorders in the offspring: a systematic review and meta-analysis". Journal of Autism and Developmental Disorders. 44 (4): 766–775. doi:10.1007/s10803-013-1928-2. PMC 4181720. PMID 24057131.
- ^ Maher GM, O'Keeffe GW, Kearney PM, Kenny LC, Dinan TG, Mattsson M, Khashan AS (August 2018). "Association of Hypertensive Disorders of Pregnancy With Risk of Neurodevelopmental Disorders in Offspring: A Systematic Review and Meta-analysis". JAMA Psychiatry. 75 (8): 809–819. doi:10.1001/jamapsychiatry.2018.0854. PMC 6143097. PMID 29874359.
- ^ Sandin S, Hultman CM, Kolevzon A, Gross R, MacCabe JH, Reichenberg A (May 2012). "Advancing maternal age is associated with increasing risk for autism: a review and meta-analysis". Journal of the American Academy of Child and Adolescent Psychiatry. 51 (5): 477–486.e1. doi:10.1016/j.jaac.2012.02.018. PMID 22525954.
- ^ Wang Y, Tang S, Xu S, Weng S, Liu Z (September 2016). "Maternal Body Mass Index and Risk of Autism Spectrum Disorders in Offspring: A Meta-analysis". Scientific Reports. 6: 34248. Bibcode:2016NatSR...634248W. doi:10.1038/srep34248. PMC 5043237. PMID 27687989.
- ^ Nieto FJ, Young TB, Lind BK, Shahar E, Samet JM, Redline S, et al. (April 2000). "Association of sleep-disordered breathing, sleep apnea, and hypertension in a large community-based study. Sleep Heart Health Study". JAMA. 283 (14): 1829–1836. doi:10.1001/jama.283.14.1829. PMID 10770144.
- ^ Muraki I, Wada H, Tanigawa T (September 2018). "Sleep apnea and type 2 diabetes". Journal of Diabetes Investigation. 9 (5): 991–997. doi:10.1111/jdi.12823. PMC 6123041. PMID 29453905. S2CID 4871197.
- ^ Punjabi NM (February 2008). "The epidemiology of adult obstructive sleep apnea". Proceedings of the American Thoracic Society. 5 (2): 136–143. doi:10.1513/pats.200709-155MG. PMC 2645248. PMID 18250205.
- ^ Bin YS, Cistulli PA, Roberts CL, Ford JB (November 2017). "Childhood Health and Educational Outcomes Associated With Maternal Sleep Apnea: A Population Record-Linkage Study". Sleep. 40 (11). doi:10.1093/sleep/zsx158. PMID 29029347.
- ^ a b Tauman R, Zuk L, Uliel-Sibony S, Ascher-Landsberg J, Katsav S, Farber M, et al. (May 2015). "The effect of maternal sleep-disordered breathing on the infant's neurodevelopment". American Journal of Obstetrics and Gynecology. 212 (5): 656.e1–656.e7. doi:10.1016/j.ajog.2015.01.001. PMID 25576821.
- ^ Perfect MM, Archbold K, Goodwin JL, Levine-Donnerstein D, Quan SF (April 2013). "Risk of behavioral and adaptive functioning difficulties in youth with previous and current sleep disordered breathing". Sleep. 36 (4): 517–525B. doi:10.5665/sleep.2536. PMC 3595180. PMID 23543901.
- ^ Murata E, Mohri I, Kato-Nishimura K, Iimura J, Ogawa M, Tachibana M, et al. (June 2017). "Evaluation of behavioral change after adenotonsillectomy for obstructive sleep apnea in children with autism spectrum disorder". Research in Developmental Disabilities. 65: 127–139. doi:10.1016/j.ridd.2017.04.012. PMID 28514706.
- ^ MacDuffie KE, Shen MD, Dager SR, Styner MA, Kim SH, Paterson S, et al. (June 2020). "Sleep Onset Problems and Subcortical Development in Infants Later Diagnosed With Autism Spectrum Disorder". The American Journal of Psychiatry. 177 (6): 518–525. doi:10.1176/appi.ajp.2019.19060666. PMC 7519575. PMID 32375538.
- ^ Santapuram, Pooja; Chen, Heidi; Weitlauf, Amy S.; Ghani, Muhammad Owais A.; Whigham, Amy S. (July 2022). "Investigating differences in symptomatology and age at diagnosis of obstructive sleep apnea in children with and without autism". International Journal of Pediatric Otorhinolaryngology. 158: 111191. doi:10.1016/j.ijporl.2022.111191. ISSN 1872-8464. PMID 35636082.
- ^ Maris, Mieke; Verhulst, Stijn; Wojciechowski, Marek; Van de Heyning, Paul; Boudewyns, An (2016-03-01). "Prevalence of Obstructive Sleep Apnea in Children with Down Syndrome". Sleep. 39 (3): 699–704. doi:10.5665/sleep.5554. ISSN 0161-8105. PMC 4763351. PMID 26612391.
- ^ Lee, Ni-Chung; Hsu, Wei-Chung; Chang, Lih-Maan; Chen, Yi-Chen; Huang, Po-Tsang; Chien, Chun-Chin; Chien, Yin-Hsiu; Chen, Chi-Ling; Hwu, Wuh-Liang; Lee, Pei-Lin (January 2020). "REM sleep and sleep apnea are associated with language function in Down syndrome children: An analysis of a community sample". Journal of the Formosan Medical Association = Taiwan Yi Zhi. 119 (1 Pt 3): 516–523. doi:10.1016/j.jfma.2019.07.015. ISSN 0929-6646. PMID 31378642.
- ^ Lord, Julia S.; Gay, Sean M.; Harper, Kathryn M.; Nikolova, Viktoriya D.; Smith, Kirsten M.; Moy, Sheryl S.; Diering, Graham H. (2022-08-29). "Early life sleep disruption potentiates lasting sex-specific changes in behavior in genetically vulnerable Shank3 heterozygous autism model mice". Molecular Autism. 13 (1): 35. doi:10.1186/s13229-022-00514-5. ISSN 2040-2392. PMC 9425965. PMID 36038911.
- ^ a b Libbey JE, Sweeten TL, McMahon WM, Fujinami RS (February 2005). "Autistic disorder and viral infections". Journal of Neurovirology (Review). 11 (1): 1–10. doi:10.1080/13550280590900553. PMID 15804954. S2CID 9962647.
- ^ Mendelsohn NJ, Schaefer GB (March 2008). "Genetic evaluation of autism". Seminars in Pediatric Neurology (Review). 15 (1): 27–31. doi:10.1016/j.spen.2008.01.005. PMID 18342258.
- ^ Meyer U, Yee BK, Feldon J (June 2007). "The neurodevelopmental impact of prenatal infections at different times of pregnancy: the earlier the worse?". The Neuroscientist (Review). 13 (3): 241–256. doi:10.1177/1073858406296401. PMID 17519367. S2CID 26096561.
- ^ Tioleco, Nina; Silberman, Anna E.; Stratigos, Katharine; Banerjee-Basu, Sharmila; Spann, Marisa N.; Whitaker, Agnes H.; Turner, J. Blake (2021). "Prenatal maternal infection and risk for autism in offspring: A meta-analysis". Autism Research. 14 (6): 1296–1316. doi:10.1002/aur.2499. ISSN 1939-3792. PMID 33720503.
- ^ Avella-Garcia CB, Julvez J, Fortuny J, Rebordosa C, García-Esteban R, Galán IR, et al. (December 2016). "Acetaminophen use in pregnancy and neurodevelopment: attention function and autism spectrum symptoms". International Journal of Epidemiology. 45 (6): 1987–1996. doi:10.1093/ije/dyw115. PMID 27353198.
- ^ a b Dufour-Rainfray D, Vourc'h P, Tourlet S, Guilloteau D, Chalon S, Andres CR (April 2011). "Fetal exposure to teratogens: evidence of genes involved in autism". Neuroscience and Biobehavioral Reviews (Review). 35 (5): 1254–1265. doi:10.1016/j.neubiorev.2010.12.013. PMID 21195109. S2CID 5180756.
- ^ Miller MT, Strömland K, Ventura L, Johansson M, Bandim JM, Gillberg C (2005). "Autism associated with conditions characterized by developmental errors in early embryogenesis: a mini review". International Journal of Developmental Neuroscience. 23 (2–3): 201–219. doi:10.1016/j.ijdevneu.2004.06.007. PMID 15749246. S2CID 14248227.
- ^ Modabbernia A, Velthorst E, Reichenberg A (2017-03-17). "Environmental risk factors for autism: an evidence-based review of systematic reviews and meta-analyses". Molecular Autism. 8 (1): 13. doi:10.1186/s13229-017-0121-4. PMC 5356236. PMID 28331572.
- ^ Samsam M, Ahangari R, Naser SA (August 2014). "Pathophysiology of autism spectrum disorders: revisiting gastrointestinal involvement and immune imbalance". World Journal of Gastroenterology (Review). 20 (29): 9942–9951. doi:10.3748/wjg.v20.i29.9942. PMC 4123375. PMID 25110424.
- ^ Román GC (November 2007). "Autism: transient in utero hypothyroxinemia related to maternal flavonoid ingestion during pregnancy and to other environmental antithyroid agents". Journal of the Neurological Sciences (Review). 262 (1–2): 15–26. doi:10.1016/j.jns.2007.06.023. PMID 17651757. S2CID 31805494.
- ^ Gardener H, Spiegelman D, Buka SL (July 2009). "Prenatal risk factors for autism: comprehensive meta-analysis". The British Journal of Psychiatry (Review, meta-analysis). 195 (1): 7–14. doi:10.1192/bjp.bp.108.051672. PMC 3712619. PMID 19567888.
- ^ Katsigianni M, Karageorgiou V, Lambrinoudaki I, Siristatidis C (December 2019). "Maternal polycystic ovarian syndrome in autism spectrum disorder: a systematic review and meta-analysis". Molecular Psychiatry. 24 (12): 1787–1797. doi:10.1038/s41380-019-0398-0. eISSN 1476-5578. PMID 30867561. S2CID 76660638.
- ^ Li YM, Ou JJ, Liu L, Zhang D, Zhao JP, Tang SY (January 2016). "Association Between Maternal Obesity and Autism Spectrum Disorder in Offspring: A Meta-analysis". Journal of Autism and Developmental Disorders. 46 (1): 95–102. doi:10.1007/s10803-015-2549-8. PMID 26254893. S2CID 26406333.
- ^ Vohr BR, Poggi Davis E, Wanke CA, Krebs NF (April 2017). "Neurodevelopment: The Impact of Nutrition and Inflammation During Preconception and Pregnancy in Low-Resource Settings". Pediatrics (Review). 139 (Suppl 1): S38–S49. doi:10.1542/peds.2016-2828F. PMID 28562247. S2CID 28637473.
- ^ Lyall K, Schmidt RJ, Hertz-Picciotto I (April 2014). "Maternal lifestyle and environmental risk factors for autism spectrum disorders". International Journal of Epidemiology. 43 (2): 443–464. doi:10.1093/ije/dyt282. PMC 3997376. PMID 24518932.
- ^ Kinney DK, Munir KM, Crowley DJ, Miller AM (October 2008). "Prenatal stress and risk for autism". Neuroscience and Biobehavioral Reviews (Review). 32 (8): 1519–1532. doi:10.1016/j.neubiorev.2008.06.004. PMC 2632594. PMID 18598714.
- ^ Rai D, Golding J, Magnusson C, Steer C, Lewis G, Dalman C (2012). "Prenatal and early life exposure to stressful life events and risk of autism spectrum disorders: population-based studies in Sweden and England". PLOS ONE. 7 (6): e38893. Bibcode:2012PLoSO...738893R. doi:10.1371/journal.pone.0038893. PMC 3374800. PMID 22719977.
- ^ Fetal testosterone and autistic traits:
- Auyeung B, Baron-Cohen S (2008). "A Role for Fetal Testosterone in Human Sex Differences". In Zimmerman AW (ed.). Autism. Humana. pp. 185–208. doi:10.1007/978-1-60327-489-0_8. ISBN 978-1-60327-488-3.
- Manson JE (October 2008). "Prenatal exposure to sex steroid hormones and behavioral/cognitive outcomes". Metabolism (Review). 57 (Suppl 2): S16–S21. doi:10.1016/j.metabol.2008.07.010. PMID 18803959.
- ^ Abramowicz JS (August 2012). "Ultrasound and autism: association, link, or coincidence?". Journal of Ultrasound in Medicine (Review). 31 (8): 1261–1269. doi:10.7863/jum.2012.31.8.1261. PMID 22837291. S2CID 36234852.
- ^ Man KK, Tong HH, Wong LY, Chan EW, Simonoff E, Wong IC (February 2015). "Exposure to selective serotonin reuptake inhibitors during pregnancy and risk of autism spectrum disorder in children: a systematic review and meta-analysis of observational studies". Neuroscience and Biobehavioral Reviews. 49: 82–89. doi:10.1016/j.neubiorev.2014.11.020. hdl:10722/207262. PMID 25498856. S2CID 8862487.
- ^ Brown HK, Hussain-Shamsy N, Lunsky Y, Dennis CE, Vigod SN (January 2017). "The Association Between Antenatal Exposure to Selective Serotonin Reuptake Inhibitors and Autism: A Systematic Review and Meta-Analysis". The Journal of Clinical Psychiatry. 78 (1): e48–e58. doi:10.4088/JCP.15r10194. PMID 28129495.
- ^ Ahlqvist, Viktor H.; Sjöqvist, Hugo; Dalman, Christina; Karlsson, Håkan; Stephansson, Olof; Johansson, Stefan; Magnusson, Cecilia; Gardner, Renee M.; Lee, Brian K. (2024). "Acetaminophen Use During Pregnancy and Children's Risk of Autism, ADHD, and Intellectual Disability". JAMA. 331 (14): 1205–1214. doi:10.1001/jama.2024.3172. PMC 11004836. PMID 38592388.
- ^ a b c Rogers CE, Lean RE, Wheelock MD, Smyser CD (December 2018). "Aberrant structural and functional connectivity and neurodevelopmental impairment in preterm children". Journal of Neurodevelopmental Disorders. 10 (1): 38. doi:10.1186/s11689-018-9253-x. eISSN 1866-1955. PMC 6291944. PMID 30541449.
- ^ Gardener H, Spiegelman D, Buka SL (August 2011). "Perinatal and neonatal risk factors for autism: a comprehensive meta-analysis". Pediatrics. 128 (2): 344–355. doi:10.1542/peds.2010-1036. PMC 3387855. PMID 21746727.
- ^ Weisskopf MG, Kioumourtzoglou MA, Roberts AL (December 2015). "Air Pollution and Autism Spectrum Disorders: Causal or Confounded?". Current Environmental Health Reports. 2 (4): 430–439. Bibcode:2015CEHR....2..430W. doi:10.1007/s40572-015-0073-9. PMC 4737505. PMID 26399256.
- ^ Flores-Pajot MC, Ofner M, Do MT, Lavigne E, Villeneuve PJ (November 2016). "Childhood autism spectrum disorders and exposure to nitrogen dioxide, and particulate matter air pollution: A review and meta-analysis". Environmental Research. 151: 763–776. Bibcode:2016ER....151..763F. doi:10.1016/j.envres.2016.07.030. PMID 27609410.
- ^ Parker W, Hornik CD, Bilbo S, Holzknecht ZE, Gentry L, Rao R, et al. (April 2017). "The role of oxidative stress, inflammation and acetaminophen exposure from birth to early childhood in the induction of autism". The Journal of International Medical Research. 45 (2): 407–438. doi:10.1177/0300060517693423. PMC 5536672. PMID 28415925.
- ^ Borchers A, Pieler T (November 2010). "Programming pluripotent precursor cells derived from Xenopus embryos to generate specific tissues and organs". Genes. 1 (3): 413–426. Bibcode:2012Entrp..14.2227S. doi:10.3390/e14112227. PMC 3966229. PMID 24710095.
- ^ Rutter M (January 2005). "Incidence of autism spectrum disorders: changes over time and their meaning". Acta Paediatrica (Review). 94 (1): 2–15. doi:10.1111/j.1651-2227.2005.tb01779.x. PMID 15858952. S2CID 79259285.
- ^ Schultz RT (2005). "Developmental deficits in social perception in autism: the role of the amygdala and fusiform face area". International Journal of Developmental Neuroscience (Review). 23 (2–3): 125–141. doi:10.1016/j.ijdevneu.2004.12.012. PMID 15749240. S2CID 17078137.
- ^ Ashwood P, Van de Water J (November 2004). "Is autism an autoimmune disease?". Autoimmunity Reviews (Review). 3 (7–8): 557–562. doi:10.1016/j.autrev.2004.07.036. PMID 15546805.
- ^ Ashwood P, Wills S, Van de Water J (July 2006). "The immune response in autism: a new frontier for autism research". Journal of Leukocyte Biology (Review). 80 (1): 1–15. doi:10.1189/jlb.1205707. PMID 16698940. S2CID 17531542.
- ^ Stigler KA, Sweeten TL, Posey DJ, McDougle CJ (2009). "Autism and immune factors: a comprehensive review". Res Autism Spectr Disord (Review). 3 (4): 840–860. doi:10.1016/j.rasd.2009.01.007.
- ^ Wills S, Cabanlit M, Bennett J, Ashwood P, Amaral D, Van de Water J (June 2007). "Autoantibodies in autism spectrum disorders (ASD)". Annals of the New York Academy of Sciences (Review). 1107 (1): 79–91. Bibcode:2007NYASA1107...79W. doi:10.1196/annals.1381.009. PMID 17804535. S2CID 24708891.
- ^ Schmitz C, Rezaie P (February 2008). "The neuropathology of autism: where do we stand?". Neuropathology and Applied Neurobiology (Review). 34 (1): 4–11. doi:10.1111/j.1365-2990.2007.00872.x. PMID 17971078. S2CID 23551620.
- ^ Wu S, Ding Y, Wu F, Li R, Xie G, Hou J, Mao P (August 2015). "Family history of autoimmune diseases is associated with an increased risk of autism in children: A systematic review and meta-analysis". Neuroscience and Biobehavioral Reviews. 55: 322–332. doi:10.1016/j.neubiorev.2015.05.004. PMID 25981892. S2CID 42029820.
- ^ Fox E, Amaral D, Van de Water J (October 2012). "Maternal and fetal antibrain antibodies in development and disease". Developmental Neurobiology (Review). 72 (10): 1327–1334. doi:10.1002/dneu.22052. PMC 3478666. PMID 22911883.
- ^ a b c Israelyan N, Margolis KG (June 2018). "Serotonin as a link between the gut-brain-microbiome axis in autism spectrum disorders". Pharmacological Research (Review). 132: 1–6. doi:10.1016/j.phrs.2018.03.020. PMC 6368356. PMID 29614380.
- ^ a b Wasilewska J, Klukowski M (2015). "Gastrointestinal symptoms and autism spectrum disorder: links and risks - a possible new overlap syndrome". Pediatric Health, Medicine and Therapeutics (Review). 6: 153–166. doi:10.2147/PHMT.S85717. PMC 5683266. PMID 29388597.
- ^ a b Rao M, Gershon MD (September 2016). "The bowel and beyond: the enteric nervous system in neurological disorders". Nature Reviews. Gastroenterology & Hepatology (Review). 13 (9): 517–528. doi:10.1038/nrgastro.2016.107. PMC 5005185. PMID 27435372.
- ^ The Editors of The Lancet (February 2010). "Retraction--Ileal-lymphoid-nodular hyperplasia, non-specific colitis, and pervasive developmental disorder in children". Lancet. 375 (9713): 445. doi:10.1016/s0140-6736(10)60175-4. PMID 20137807. S2CID 26364726.
- ^ Bezawada N, Phang TH, Hold GL, Hansen R (2020). "Autism Spectrum Disorder and the Gut Microbiota in Children: A Systematic Review". Annals of Nutrition & Metabolism. 76 (1): 16–29. doi:10.1159/000505363. eISSN 1421-9697. PMID 31982866. S2CID 210922352.
- ^ Almeida C, Oliveira R, Soares R, Barata P (2020). "Influence of gut microbiota dysbiosis on brain function: a systematic review". Porto Biomedical Journal. 5 (2): 1. doi:10.1097/j.pbj.0000000000000059. eISSN 2444-8672. PMC 7722401. PMID 33299942.
- ^ a b c d Azhari A, Azizan F, Esposito G (July 2019). "A systematic review of gut-immune-brain mechanisms in Autism Spectrum Disorder". Developmental Psychobiology (Systematic Review). 61 (5): 752–771. doi:10.1002/dev.21803. hdl:10220/49107. PMID 30523646. S2CID 54523742.
- ^ Johnson TW (2006). "Dietary considerations in autism: identifying a reasonable approach". Top Clin Nutr. 21 (3): 212–225. doi:10.1097/00008486-200607000-00008. S2CID 76326593.
- ^ Krishnaswami S, McPheeters ML, Veenstra-Vanderweele J (May 2011). "A systematic review of secretin for children with autism spectrum disorders". Pediatrics (Review). 127 (5): e1322–e1325. doi:10.1542/peds.2011-0428. PMC 3387870. PMID 21464196.
- ^ Panksepp J (1979). "A neurochemical theory of autism". Trends in Neurosciences. 2: 174–177. doi:10.1016/0166-2236(79)90071-7. S2CID 54373822.
- ^ a b c d Millward C, Ferriter M, Calver S, Connell-Jones G (April 2008). "Gluten- and casein-free diets for autistic spectrum disorder". The Cochrane Database of Systematic Reviews (Review) (2): CD003498. doi:10.1002/14651858.CD003498.pub3. PMC 4164915. PMID 18425890.
- ^ Shattock P, Whiteley P (April 2002). "Biochemical aspects in autism spectrum disorders: updating the opioid-excess theory and presenting new opportunities for biomedical intervention". Expert Opinion on Therapeutic Targets (Review). 6 (2): 175–183. doi:10.1517/14728222.6.2.175. PMID 12223079. S2CID 40904799.
- ^ Christison GW, Ivany K (April 2006). "Elimination diets in autism spectrum disorders: any wheat amidst the chaff?". Journal of Developmental and Behavioral Pediatrics. 27 (2 Suppl): S162–S171. doi:10.1097/00004703-200604002-00015. PMID 16685183.
- ^ a b Aranburu E, Matias S, Simón E, Larretxi I, Martínez O, Bustamante MÁ, et al. (May 2021). "Gluten and FODMAPs Relationship with Mental Disorders: Systematic Review". Nutrients. 13 (6): 1894. doi:10.3390/nu13061894. PMC 8228761. PMID 34072914.
- ^ a b c d e f g h i j k Modabbernia A, Velthorst E, Reichenberg A (2017-03-17). "Environmental risk factors for autism: an evidence-based review of systematic reviews and meta-analyses". Molecular Autism. 8 (1): 13. doi:10.1186/s13229-017-0121-4. PMC 5356236. PMID 28331572.
- ^ Rossignol DA, Frye RE (November 2021). "Cerebral Folate Deficiency, Folate Receptor Alpha Autoantibodies and Leucovorin (Folinic Acid) Treatment in Autism Spectrum Disorders: A Systematic Review and Meta-Analysis". Journal of Personalized Medicine. 11 (11): 1141. doi:10.3390/jpm11111141. PMC 8622150. PMID 34834493.
- ^ a b c Jafari Mohammadabadi H, Rahmatian A, Sayehmiri F, Rafiei M (2020-09-21). "The Relationship Between the Level of Copper, Lead, Mercury and Autism Disorders: A Meta-Analysis". Pediatric Health, Medicine and Therapeutics. 11: 369–378. doi:10.2147/PHMT.S210042. PMC 7519826. PMID 33061742.
- ^ Islam GM, Rahman MM, Hasan MI, Tadesse AW, Hamadani JD, Hamer DH (March 2022). "Hair, serum and urine chromium levels in children with cognitive defects: A systematic review and meta-analysis of case control studies". Chemosphere. 291 (Pt 2): 133017. Bibcode:2022Chmsp.29133017I. doi:10.1016/j.chemosphere.2021.133017. eISSN 1879-1298. PMC 8792285. PMID 34813844.
- ^ Zafeiriou DI, Ververi A, Vargiami E (June 2007). "Childhood autism and associated comorbidities". Brain & Development (Review). 29 (5): 257–272. doi:10.1016/j.braindev.2006.09.003. PMID 17084999. S2CID 16386209.
- ^ Mehler MF, Purpura DP (March 2009). "Autism, fever, epigenetics and the locus coeruleus". Brain Research Reviews (Review). 59 (2): 388–392. doi:10.1016/j.brainresrev.2008.11.001. PMC 2668953. PMID 19059284. For a lay summary, see Kluger J (2009-04-07). "Why Fever Helps Autism: A New Theory". Time. Archived from the original on 2013-03-28.
- ^ Markkanen E, Meyer U, Dianov GL (June 2016). "DNA Damage and Repair in Schizophrenia and Autism: Implications for Cancer Comorbidity and Beyond". International Journal of Molecular Sciences. 17 (6): 856. doi:10.3390/ijms17060856. PMC 4926390. PMID 27258260.
- ^ a b c d Bjørklund G, Meguid NA, El-Bana MA, Tinkov AA, Saad K, Dadar M, et al. (May 2020). "Oxidative Stress in Autism Spectrum Disorder". Molecular Neurobiology. 57 (5): 2314–2332. doi:10.1007/s12035-019-01742-2. PMID 32026227. S2CID 255484540.
- ^ Kern JK, Jones AM (2006). "Evidence of toxicity, oxidative stress, and neuronal insult in autism". Journal of Toxicology and Environmental Health Part B: Critical Reviews (Review). 9 (6): 485–499. Bibcode:2006JTEHB...9..485K. doi:10.1080/10937400600882079. PMID 17090484. S2CID 1096750.
- ^ Ghanizadeh A, Akhondzadeh S, Hormozi M, Makarem A, Abotorabi-Zarchi M, Firoozabadi A (2012). "Glutathione-related factors and oxidative stress in autism, a review". Current Medicinal Chemistry (Review). 19 (23): 4000–4005. doi:10.2174/092986712802002572. PMID 22708999.
- ^ Villagonzalo KA, Dodd S, Dean O, Gray K, Tonge B, Berk M (December 2010). "Oxidative pathways as a drug target for the treatment of autism". Expert Opinion on Therapeutic Targets (Review). 14 (12): 1301–1310. doi:10.1517/14728222.2010.528394. PMID 20954799. S2CID 44864562.
- ^ a b c d Moore I, Morgan G, Welham A, Russell G (November 2022). "The intersection of autism and gender in the negotiation of identity: A systematic review and metasynthesis". Feminism & Psychology. 32 (4): 421–442. doi:10.1177/09593535221074806. ISSN 0959-3535. S2CID 246893906.
- ^ Hacking I (1999). The Social Construction of What?. Harvard University Press. pp. 114–123. ISBN 0-674-00412-4.
- ^ Nadesan MH (2005). "The dialectics of autism: theorizing autism, performing autism, remediating autism, and resisting autism". Constructing Autism: Unravelling the 'Truth' and Understanding the Social. Routledge. pp. 179–213. ISBN 0-415-32181-6.
- ^ Waterhouse L (2013). Rethinking Autism: Variation and Complexity. Academic Press. p. 24. ISBN 978-0-12-415961-7.
Although autism spectrum disorder has not been proven to exist either as a set of meaningful subgroups, or as the expression of a unifying deficit or causal pattern, nonetheless, autism appears to have been unified as a real entity in public opinion... Some researchers have argued that, over time, autism has been transformed from a hypothesis to an assumed reality. This transformation is called reification. Reification is the conversion of a theorized entity into something assumed and believed to be real... the intense public discussion of autism, the long history of autism in the diagnostic manuals of the American Psychiatric Association, and the long history of autism research are in full view, and they all have made autism seem more concrete and less hypothetical.
- ^ Spikins P (March 27, 2017). "How our autistic ancestors played an important role in human evolution". The Conversation.
- ^ Spikins P (March 6, 2013). "The Stone Age Origins of Autism". In Fitzgerald M (ed.). Recent Advances in Autism Spectrum Disorders - Volume II.
- ^ Lomelin DE (2010). "An Examination of Autism Spectrum Disorders in Relation to Human Evolution and Life History Theory". Nebraska Anthropologist. 57.
- ^ Reser JE (May 2011). "Conceptualizing the autism spectrum in terms of natural selection and behavioral ecology: the solitary forager hypothesis". Evolutionary Psychology. 9 (2): 207–238. doi:10.1177/147470491100900209. PMC 10480880. PMID 22947969. S2CID 25378900.
- ^ "Autism may have had advantages in humans' hunter-gatherer past, researcher believes". ScienceDaily. June 3, 2011.
- ^ Rudacille D (8 July 2011). "Lonely hunters". Spectrum.
- ^ Spikins P, Wright B, Hodgson D (1 October 2016). "Are there alternative adaptive strategies to human pro-sociality? The role of collaborative morality in the emergence of personality variation and autistic traits". Time and Mind. 9 (4): 289–313. doi:10.1080/1751696X.2016.1244949. ISSN 1751-696X. S2CID 151820168.
- ^ Nesse, Randolph M. (2019). "14. Minds Unbalanced on Fitness Cliffs". Good Reasons for Bad Feelings: Insights from the Frontier of Evolutionary Psychiatry. New York: Dutton. pp. 245–261. ISBN 978-1101985663.
- ^ Nesse, Randolph M. (2016) [2005]. "43. Evolutionary Psychology and Mental Health". In Buss, David M. (ed.). The Handbook of Evolutionary Psychology, Volume 2: Integrations (2nd ed.). Hoboken, NJ: Wiley. pp. 1018–1019. ISBN 978-1118755808.
- ^ Nesse, Randolph M. (March 4, 2019). "The Puzzle of the Unbalanced Mind". Psychology Today. Retrieved October 13, 2024.
- ^ Fu, Q.; Hajdinjak, M.; Moldovan, O. T.; et al. (2015). "An early modern human from Romania with a recent Neanderthal ancestor". Nature. 524 (7564): 216–219. Bibcode:2015Natur.524..216F. doi:10.1038/nature14558. PMC 4537386. PMID 26098372.
- ^ Rehnström, Karola. "Genetic Heterogeneity in Autism Spectrum Disorders in a Population Isolate" (PDF). Julkari. National Institute for Health and Welfare (Finland). Retrieved 24 March 2024.
- ^ a b c Green, R. E.; Krause, J.; Briggs, A. W.; et al. (2010). "A draft sequence of the Neandertal genome". Science. 328 (5979): 710–722. Bibcode:2010Sci...328..710G. doi:10.1126/science.1188021. PMC 5100745. PMID 20448178.
- ^ Sankararaman, S.; Patterson, N.; Li, H.; Pääbo, S.; Reich, D; Akey, J. M. (2012). "The date of interbreeding between Neandertals and modern humans". PLOS Genetics. 8 (10): e1002947. arXiv:1208.2238. Bibcode:2012arXiv1208.2238S. doi:10.1371/journal.pgen.1002947. PMC 3464203. PMID 23055938.
- ^ Yang, M. A.; Malaspinas, A. S.; Durand, E. Y.; Slatkin, M. (2012). "Ancient structure in Africa unlikely to explain Neanderthal and non-African genetic similarity". Molecular Biology and Evolution. 29 (10): 2, 987–2, 995. doi:10.1093/molbev/mss117. PMC 3457770. PMID 22513287.
- ^ Sánchez-Quinto, F.; Botigué, L. R.; Civit, S.; Arenas, C.; Ávila-Arcos, M. C.; Bustamante, C. D.; Comas, D.; Lalueza-Fox, C.; Caramelli, D. (2012). "North African populations carry the signature of admixture with Neandertals". PLOS ONE. 7 (10): e47765. Bibcode:2012PLoSO...747765S. doi:10.1371/journal.pone.0047765. PMC 3474783. PMID 23082212.
- ^ Sankararaman, S.; Mallick, S.; Dannemann, M.; Prüfer, K.; Kelso, J.; Pääbo, S.; Patterson, N.; Reich, D. (2014). "The genomic landscape of Neanderthal ancestry in present-day humans". Nature. 507 (7492): 354–357. Bibcode:2014Natur.507..354S. doi:10.1038/nature12961. PMC 4072735. PMID 24476815.
- ^ Yotova, V.; Lefebvre, J.-F.; Moreau, C.; et al. (2011). "An X-linked haplotype of Neandertal origin is present among all non-African populations". Molecular Biology and Evolution. 28 (7): 1957–1962. doi:10.1093/molbev/msr024. PMID 21266489.
- ^ Fu, Q.; Li, H.; Moorjani, P.; et al. (2014). "Genome sequence of a 45,000-year-old modern human from western Siberia". Nature. 514 (7523): 445–449. Bibcode:2014Natur.514..445F. doi:10.1038/nature13810. PMC 4753769. PMID 25341783.
- ^ a b c Chen, L.; Wolf, A. B.; Fu, W.; Akey, J. M. (2020). "Identifying and Interpreting Apparent Neanderthal Ancestry in African Individuals". Cell. 180 (4): 677–687.e16. doi:10.1016/j.cell.2020.01.012. PMID 32004458. S2CID 210955842.
- ^ Lohse, K.; Frantz, L. A. F. (2013). "Maximum likelihood evidence for Neandertal admixture in Eurasian populations from three genomes". Populations and Evolution. 1307: 8263. arXiv:1307.8263. Bibcode:2013arXiv1307.8263L.
- ^ Prüfer, K.; de Filippo, C.; Grote, S.; Mafessoni, F.; Korlević, P.; Hajdinjak, M.; et al. (2017). "A high-coverage Neandertal genome from Vindija Cave in Croatia". Science. 358 (6363): 655–658. Bibcode:2017Sci...358..655P. doi:10.1126/science.aao1887. PMC 6185897. PMID 28982794.
- ^ a b c Reich 2018.
- ^ Pääbo, S. (2015). "The diverse origins of the human gene pool". Nature Reviews Genetics. 16 (6): 313–314. doi:10.1038/nrg3954. PMID 25982166. S2CID 5628263.
- ^ Enard, D.; Petrov, D. A. (2018). "Evidence that RNA viruses drove of adaptive introgression between Neanderthals and modern humans". Cell. 175 (2): 360–371. doi:10.1016/j.cell.2018.08.034. PMC 6176737. PMID 30290142.
- ^ Vernot, B.; Akey, J. M. (2014). "Resurrecting surviving Neandertal lineages from modern human genomes". Science. 343 (6174): 1017–1021. Bibcode:2014Sci...343.1017V. doi:10.1126/science.1245938. PMID 24476670. S2CID 23003860.
- ^ Juric, I.; Aeschbacher, S.; Coop, G. (2016). "The strength of selection against Neanderthal introgression". PLOS Genetics. 12 (11): e1006340. doi:10.1371/journal.pgen.1006340. PMC 5100956. PMID 27824859.
- ^ Taskent, R. O.; Alioglu, N. D.; Fer, E.; et al. (2017). "Variation and functional impact of Neanderthal ancestry in Western Asia". Genome Biology and Evolution. 9 (12): 3516–3624. doi:10.1093/gbe/evx216. PMC 5751057. PMID 29040546.
- ^ Zorina-Lichtenwalter, K.; Lichtenwalter, R. N.; Zaykin, D. V.; et al. (2019). "A study in scarlet: MC1R as the main predictor of red hair and exemplar of the flip-flop effect". Human Molecular Genetics. 28 (12): 2093–2106. doi:10.1093/hmg/ddz018. PMC 6548228. PMID 30657907.
- ^ Ding, Q.; Hu, Y.; Xu, S.; Wang, C.-C.; Li, H.; Zhang, R.; Yan, S.; Wang, J.; Jin, L. (2014). "Neanderthal origin of the haplotypes carrying the functional variant Val92Met in the MC1R in modern humans". Molecular Biology and Evolution. 31 (8): 1994–2003. doi:10.1093/molbev/msu180. PMID 24916031. "We further discovered that all of the putative Neanderthal introgressive haplotypes carry the Val92Met variant, a loss-of-function variant in MC1R that is associated with multiple dermatological traits including skin color and photoaging. Frequency of this Neanderthal introgression is low in Europeans (~5%), moderate in continental East Asians (~30%), and high in Taiwanese aborigines (60–70%)."
- ^ Ségurel, L.; Quintana-Murci, L. (2014). "Preserving immune diversity through ancient inheritance and admixture". Current Opinion in Immunology. 30: 79–84. doi:10.1016/j.coi.2014.08.002. PMID 25190608.
- ^ a b Mendez, F. L.; Watkins, J. C.; Hammer, M. F. (2013). "Neandertal origin of genetic variation at the cluster of OAS immunity genes" (PDF). Molecular Biology and Evolution. 30 (4): 798–801. doi:10.1093/molbev/mst004. PMID 23315957. S2CID 2839679. Archived from the original (PDF) on 2019-02-23.
- ^ a b Mendez, F. L.; Watkins, J. C.; Hammer, M. F. (2012). "A haplotype at STAT2 introgressed from Neanderthals and serves as a candidate of positive selection in Papua New Guinea". American Journal of Human Genetics. 91 (2): 265–274. doi:10.1016/j.ajhg.2012.06.015. PMC 3415544. PMID 22883142.
- ^ a b Dannemann, M.; Andrés, A. A.; Kelso, J. (2016). "Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human toll-like receptors". American Journal of Human Genetics. 98 (1): 22–33. doi:10.1016/j.ajhg.2015.11.015. PMC 4716682. PMID 26748514.
- ^ a b Nédélec, Y.; Sanz, J.; Baharian, G.; et al. (2016). "Genetic ancestry and natural selection drive population differences in immune responses to pathogens". Cell. 167 (3): 657–669. doi:10.1016/j.cell.2016.09.025. PMID 27768889.
- ^ Gregory, M. D.; Kippenhan, J. S.; Eisenberg, D. P.; et al. (2017). "Neanderthal-derived genetic variation shapes modern human cranium and brain". Scientific Reports. 7 (1): 6308. Bibcode:2017NatSR...7.6308G. doi:10.1038/s41598-017-06587-0. PMC 5524936. PMID 28740249.
- ^ Dolgova, O.; Lao, O. (2018). "Evolutionary and medical consequences of archaic introgression into modern human genomes". Genes. 9 (7): 358. doi:10.3390/genes9070358. PMC 6070777. PMID 30022013.
- ^ "Human-Neanderthal Gene Variance is Involved in Autism". Neuroscience News. 4 August 2016. Retrieved 24 March 2024.
- ^ McCarthy, Michael (4 August 2016). "Human-Neanderthal gene variance is involved in autism". Medical Express. Retrieved 24 March 2024.
- ^ Gregory, Michael D.; Kippenhan, J. Shane; Eisenberg, Daniel P.; Kohn, Philip D.; Dickinson, Dwight; Mattay, Venkata S.; Chen, Qiang; Weinberger, Daniel R.; Saad, Ziad S.; Berman, Karen F. (24 July 2017). "Neanderthal-Derived Genetic Variation Shapes Modern Human Cranium and Brain". Scientific Reports. 7 (1): 6308. Bibcode:2017NatSR...7.6308G. doi:10.1038/s41598-017-06587-0. PMC 5524936. PMID 28740249.
- ^ Mozzi, Alessandra; Forni, Diego; Cagliani, Rachele; Pozzoli, Uberto; Clerici, Mario; Sironi, Manuela (21 July 2017). "Distinct selective forces and Neanderthal introgression shaped genetic diversity at genes involved in neurodevelopmental disorders". Scientific Reports. 7 (6116): 6116. Bibcode:2017NatSR...7.6116M. doi:10.1038/s41598-017-06440-4. hdl:2434/554557. PMC 5522412. PMID 28733602.
- ^ Prakash, Anil; Banerjee, Moinak (13 May 2021). "Genomic selection signatures in autism spectrum disorder identifies cognitive genomic tradeoff and its relevance in paradoxical phenotypes of deficits versus potentialities". Scientific Reports. 11 (10245): 10245. Bibcode:2021NatSR..1110245P. doi:10.1038/s41598-021-89798-w. PMC 8119484. PMID 33986442.
- ^ Bettelheim B (1967). The Empty Fortress: Infantile Autism and the Birth of the Self. Free Press. ISBN 0-02-903140-0.
- ^ Kanner L (1943). "Autistic disturbances of affective contact". Nerv Child. 2: 217–250. Reprinted in Kanner L (1968). "Autistic disturbances of affective contact". Acta Paedopsychiatrica. 35 (4): 100–136. PMID 4880460.
- ^ Kanner L (July 1949). "Problems of nosology and psychodynamics of early infantile autism". The American Journal of Orthopsychiatry. 19 (3): 416–426. doi:10.1111/j.1939-0025.1949.tb05441.x. PMID 18146742.
- ^ Gardner M (2000). "The brutality of Dr. Bettelheim". Skeptical Inquirer. 24 (6): 12–14.
- ^ King's College London (April 24, 2023). "Peering Into the Womb: Fetal Brain Scans Reveal Autism Clues". SciTechDaily.
- ^ Fombonne E, Zakarian R, Bennett A, Meng L, McLean-Heywood D (July 2006). "Pervasive developmental disorders in Montreal, Quebec, Canada: prevalence and links with immunizations". Pediatrics. 118 (1): e139–e150. doi:10.1542/peds.2005-2993. PMID 16818529. S2CID 17981294.
- ^ Gross L (May 2009). "A broken trust: lessons from the vaccine--autism wars". PLOS Biology. 7 (5): e1000114. doi:10.1371/journal.pbio.1000114. PMC 2682483. PMID 19478850.
- ^ Taylor LE, Swerdfeger AL, Eslick GD (June 2014). "Vaccines are not associated with autism: an evidence-based meta-analysis of case-control and cohort studies". Vaccine. 32 (29): 3623–3629. doi:10.1016/j.vaccine.2014.04.085. PMID 24814559.
- ^ Hilton S, Petticrew M, Hunt K (May 2006). "'Combined vaccines are like a sudden onslaught to the body's immune system': parental concerns about vaccine 'overload' and 'immune-vulnerability'". Vaccine. 24 (20): 4321–4327. doi:10.1016/j.vaccine.2006.03.003. PMID 16581162.
- ^ Gerber JS, Offit PA (February 2009). "Vaccines and autism: a tale of shifting hypotheses". Clinical Infectious Diseases (Review). 48 (4): 456–461. doi:10.1086/596476. PMC 2908388. PMID 19128068. For a lay summary,
- ^ Paul R (June 2009). "Parents ask: Am I risking autism if I vaccinate my children?". Journal of Autism and Developmental Disorders. 39 (6): 962–963. doi:10.1007/s10803-009-0739-y. PMID 19363650. S2CID 34467853.
- ^ Foster CA, Ortiz SM (2017). "Vaccines, Autism, and the Promotion of Irrelevant Research: A Science-Pseudoscience Analysis". Skeptical Inquirer. 41 (3): 44–48. Archived from the original on 2018-10-06. Retrieved 6 October 2018.
- ^ a b c The Editors Of The Lancet (February 2010). "Retraction--Ileal-lymphoid-nodular hyperplasia, non-specific colitis, and pervasive developmental disorder in children". Lancet. 375 (9713): 445. doi:10.1016/S0140-6736(10)60175-4. PMID 20137807. S2CID 26364726. For a lay summary, see Triggle N (2010-02-02). "Lancet accepts MMR study 'false'". BBC News.
- ^ Wakefield AJ, Murch SH, Anthony A, Linnell J, Casson DM, Malik M, et al. (February 1998). "Ileal-lymphoid-nodular hyperplasia, non-specific colitis, and pervasive developmental disorder in children". Lancet. 351 (9103): 637–641. doi:10.1016/S0140-6736(97)11096-0. PMID 9500320. S2CID 439791. (Retracted, see doi:10.1016/S0140-6736(10)60175-7)
- ^ Murch SH, Anthony A, Casson DH, Malik M, Berelowitz M, Dhillon AP, et al. (March 2004). "Retraction of an interpretation". Lancet. 363 (9411): 750. doi:10.1016/S0140-6736(04)15715-2. PMID 15016483. S2CID 5128036.
- ^ Deer B (2008-11-02). "The MMR-autism crisis – our story so far". Retrieved 2008-12-06.
- ^ a b "Measles, mumps, and rubella (MMR) vaccine". Centers for Disease Control and Prevention. 2008-12-23. Retrieved 2009-02-14.
- ^ a b "Immunization safety review: vaccines and autism". Institute of Medicine, National Academy of Sciences. 2004. Archived from the original on 2007-06-23. Retrieved 2007-06-13.
- ^ "MMR the facts". National Health Service. Archived from the original on 2007-06-15. Retrieved 2007-06-13.
- ^ Godlee F, Smith J, Marcovitch H (January 2011). "Wakefield's article linking MMR vaccine and autism was fraudulent". BMJ. 342: c7452. doi:10.1136/bmj.c7452. PMID 21209060. S2CID 43640126.
- ^ Deer B (January 2011). "How the case against the MMR vaccine was fixed". BMJ. 342: c5347. doi:10.1136/bmj.c5347. PMID 21209059. S2CID 46683674.
- ^ "Study linking vaccine to autism was fraud". NPR. Associated Press. 2011-01-05. Archived from the original on 2011-01-07. Retrieved 2011-01-06.
- ^ "Retracted autism study an 'elaborate fraud,' British journal finds". Atlanta. 2011-01-06. Retrieved 2011-01-06.
- ^ "Vaccines, blood and biologics: thimerosal in vaccines". US Food and Drug Administration. 2012. Retrieved October 24, 2013.
- ^ Gorski D (7 January 2008). "Mercury in vaccines as a cause of autism and autism spectrum disorders (ASDs): A failed hypothesis". Science-Based Medicine.
- ^ American Medical Association (2004-05-18). "AMA Welcomes New IOM Report Rejecting Link Between Vaccines and Autism". Archived from the original on 2012-10-06. Retrieved 2007-07-23.
- ^ American Academy of Pediatrics (2004-05-18). "What Parents Should Know About Thimerosal". Archived from the original on 2007-07-08. Retrieved 2007-07-23.
- ^ Kurt TL (December 2006). "ACMT position statement: the Iom report on thimerosal and autism" (PDF). Journal of Medical Toxicology. 2 (4): 170–171. doi:10.1007/BF03161188. PMC 3550071. PMID 18072140. Archived from the original (PDF) on 2008-02-29.
- ^ Infectious Diseases and Immunization Committee, Canadian Paediatric Society (May 2007). "Autistic spectrum disorder: No causal relationship with vaccines". Paediatrics & Child Health. 12 (5): 393–398. PMC 2528717. PMID 19030398. Archived from the original on 2008-12-02. Retrieved 2008-10-17. Also published (2007) in "Autistic spectrum disorder: No causal relationship with vaccines". The Canadian Journal of Infectious Diseases & Medical Microbiology. 18 (3): 177–179. May 2007. doi:10.1155/2007/267957. PMC 2533550. PMID 18923720..
- ^ "Thimerosal in vaccines". Center for Biologics Evaluation and Research, U.S. Food and Drug Administration. 2007-09-06. Retrieved 2007-10-01.
- ^ World Health Organization (2006). "Questions and answers about autism spectrum disorders (ASD)". Retrieved 2014-11-02.
- ^ National Advisory Committee on Immunization (July 2007). "Thimerosal: updated statement. An Advisory Committee Statement (ACS)". Canada Communicable Disease Report. 33 (ACS-6): 1–13. PMID 17663033.
- ^ European Medicines Agency (2004-03-24). "EMEA Public Statement on Thiomersal in Vaccines for Human Use" (PDF). Archived from the original (PDF) on 2007-06-10. Retrieved 2007-07-22.
Bibliography
edit- Reich, D. (2018). "Encounters with Neanderthals". Who we are and how we got here: ancient DNA and the new science of the human past. Oxford University Press. ISBN 978-0-19-882125-0.