TM6SF2 is the Transmembrane 6 superfamily 2 human gene which codes for a protein by the same name. This gene is otherwise called KIAA1926.[5] Its exact function is currently unknown.
| TM6SF2 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | TM6SF2, transmembrane 6 superfamily member 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 606563; MGI: 1933210; HomoloGene: 77694; GeneCards: TM6SF2; OMA:TM6SF2 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Location
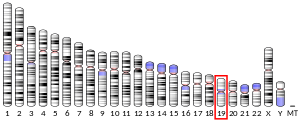
editTM6SF2 is located on chromosome 19 precisely at locus 19p13.3-p12. It is flanked by SUGP1 (a SURP and G-Patch Domain-Containing protein thought to play a role in pre-mRNA splicing [5]) and HAPLN4 (a hyaluronan and proteoglycan link protein 4 that binds to hyaluronic acid and may be involved in formation of the extracellular matrix [5]) genes upstream and downstream respectively.[6]
Evolutionary aspects
editOrthologs
editTM6SF2 is a moderately conserved gene. There exist orthologs in several phyla as far diverged as invertebrates. 82 organisms have been identified as having orthologs of this gene. The most distant orthologs of TM6SF2 are in zebra fish (Danio rerio) and the deer tick (Ixodes scapularis).[6] Below is a summary table of some of the gene orthologs obtained from the NCBI database.
| Scientific Name | Common Name | Divergence date (MYA) | NCBI [6] accession number | Sequence Length | Percent Identity | Percent Similarity |
|---|---|---|---|---|---|---|
| Homo sapiens | Human | 0 | NP_001001524.2 | 377 | 100 | 100 |
| Pan troglodytes | Chimpanzee | 6.3 | XP_001140342.2 | 377 | 99 | 99 |
| Mus musculus | Mouse | 92.3 | XP_003125904 | 378 | 79 | 87 |
| Ceratotherium simum simum | Southern white rhinoceros | 94.2 | XP_004422975.1 | 376 | 89 | 92 |
| Capra hircus | Goat | 94.2 | XP_005682141.1 | 343 | 89 | 86 |
| Myotis davidii | Mouse-eared bat | 94.2 | XP_006778388.1 | 338 | 86 | 91 |
| Mustela putorius furo | Domestic ferret | 94.2 | XP_004760922.1 | 376 | 84 | 89 |
| Vicugna pacos | Alpaca | 94.2 | XP_006199087.1 | 376 | 84 | 89 |
| Canis lupus familiaris | Dog | 94.2 | XP_852125.1 | 376 | 83 | 89 |
| Orcinus orca | Killer whale | 94.2 | XP_004277546.1 | 376 | 82 | 88 |
| Bos taurus | Cow | 94.2 | XP_005208509.1 | 376 | 74 | 80 |
| Loxodonta africana | African savanna elephant | 98.7 | XP_003413566.1 | 377 | 90 | 93 |
| Alligator mississipiens | American alligator | 296 | XP_006271093.1 | 346 | 67 | 79 |
| Ophiophagus hannah | King cobra | 296 | ETE70999 | 292 | 25.3 | ? |
| Gallus gallus | Chicken | 296 | XP_423447.3 | 374 | 62 | 74 |
| Falco peregrinus | Peregrine falcon | 296 | XP_005244205.1 | 376 | 59 | 73 |
| Xenopus tropicalis | Western-clawed frog | 371.2 | XP_004760922.1 | 375 | 58 | 74 |
| Danio rerio | Zebrafish | 400.1 | NP_001074130 | 374 | 44.3 | ? |
| Latimeria chalumnae | Coelocanth | 414.9 | XP_005989673.1 | 327 | 63 | 75 |
| Ixodes scapularis | Deer tick | 782.7 | XP_002406440.1 | 113 | 45.1 | ? |
Paralogs
editTM6SF1 has been identified as a paralog of TM6SF2 in humans [6] about which little is known.
Homologous domains
editThe domain of unknown function DUF2781 is highly conserved across homologs. DUF2781 belongs to the pfam10914 family which comprises uncharacterized eukaryotic proteins, some of which are membrane proteins [6]
mRNA
editThe RNA product is 1483 base pairs long and is spliced alternatively to yield seven different isoforms (alternative mRNAs a - f with form a being the most abundant) with varying combinations of the 10 identified exons.[7] The microRNA miR-1343 binds to a 3’ UTR site called 7mer-m8 (as predicted by TargetScan[8]).
Folding patterns
editThe 5' and 3' UTR regions of the mRNA show some stem loop formation for stability. Much of this chemistry appears to be taking place in the 5' region which has three stem loops compared to the 3' region with only one.[9]
Exons and introns
editThere are ten different exons and the ones expressed depend on how alternative splicing proceeds. There are four alternative polyadenylation sites present.[7]
Promoter region
editThe promoter for this gene is upstream and spans bases 19383923 to 19384700 (778 bp long) on the minus strand of chromosome 19. There exist several transcription factors capable of binding to this promoter region including cAMP responsive element binding protein, SMAD3, KLF3, EGR1, SOX/SRY, PAX2/PAX5[10] and two SNP regions have been identified as well.[11] The transcription factors predicted to bind the TM6SF2 promoter suggest this protein functions in growth and tumor regulation as well as sex determination to a lesser extent.
Protein
editThe TM6SF2 protein contains 377 amino acids and is 42,554 Da large with an isoelectric point of about 7.7.[12]
Domains and motifs
editThere is a domain of unknown function, DUF2781 ( pfam10914 family) spanning amino acids 218 to 359 in the C-terminus of the protein.[6] There are nine transmembrane regions in this protein. The first one contains the signal peptide which is eventually cleaved following protein localization to the ER. A terminal KHHQ sequence is an endoplasmic reticulum retention signal.[13]
Secondary structure
editSeveral alpha helices and beta strands are formed by the mature protein with as many as thirteen helices (including transmembrane helices) and fifteen beta sheets predicted.[14]
3° and 4° structure
editThe protein side groups in this protein do not necessarily interact in a manner to form tertiary and quaternary structures. The cysteines present are not predicted to form stable disulfide bonds.[15]
Post-translational modifications
editTwo main post-translational modifications occur; phosphorylation at tyrosine, serine and tryptophan sites and two low probability sumoylation sites.[16]
Expression patterns
editIn humans, TM6SF2 expression has been documented in the adult stage only specifically in the intestine and liver in moderate amounts as well as embryonic tissue and ovary at low levels. Other sources indicate expression in brain, lung, testis, stomach, heart, colon, kidney and adipose tissue.[17]
Protein subcellular localization studies with confocal microscopy demonstrated that TM6SF2 is localized in the endoplasmic reticulum and the ER-Golgi intermediate compartment of human liver cells.[18]
Protein interactions
editNo known protein-protein interactions have been established thus far.[19][20][21]
Clinical significance
editIn a study that used pre-made kits to predict cardiac allograft rejection using peripheral blood only, graft rejection was associated with decreased levels of TM6SF2 expression, alongside other genes.[22]
A variant TM6SF2 gene causes susceptibility to nonalcoholic fatty liver disease due to impaired very low density lipoprotein (VLDL) production.[23]
TM6SF2 inhibition was associated with reduced secretion of TG-rich lipoproteins (TRLs) and increased cellular TG concentration and lipid droplet content, whereas TM6SF2 overexpression reduced liver cell steatosis. TM6SF2 is a regulator of liver fat metabolism with opposing effects on the secretion of TRLs and hepatic lipid droplet content.[18]
References
edit- ^ a b c GRCh38: Ensembl release 89: ENSG00000213996 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000036151 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c GeneCards® The Human Database Compendium website https://www.genecards.org/
- ^ a b c d e f National Center for Biotechnology Information, U.S. National Library of Medicine website https://www.ncbi.nlm.nih.gov/
- ^ a b National Center for Biotechnology Information, U.S. National Library of Medicine website. AceView program.https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/
- ^ Agarwal V, Bell GW, Nam JW, Bartel DP (August 2015). "Predicting effective microRNA target sites in mammalian mRNAs". eLife. 4: e05005. doi:10.7554/eLife.05005. PMC 4532895. PMID 26267216.
- ^ mfold web server: 1995-2014, Michael Zuker & Nick Markham, © Rensselaer Polytechnic Institute. Hosted by The RNA Institute, College of Arts and Sciences, State University of New York at Albany.http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form
- ^ Genomatix Gene2Promoter program. http://www.genomatix.de/ Archived 2021-12-02 at the Wayback Machine
- ^ National Center for Biotechnology Information, U.S. National Library of Medicine website. SNP:GeneView program.https://www.ncbi.nlm.nih.gov/SNP/
- ^ San Diego Super Computer (SDSC) Biology Workbench online program. SAPS protein assessment program.http://workbench.sdsc.edu/
- ^ Petersen TN, Brunak S, von Heijne G, Nielsen H (September 2011). "SignalP 4.0: discriminating signal peptides from transmembrane regions". Nature Methods. 8 (10): 785–6. doi:10.1038/nmeth.1701. PMID 21959131. S2CID 16509924.
- ^ San Diego Super Computer (SDSC) Biology Workbench online program. CHOFAS program.
- ^ DISULFIND program: Ceroni, A. Passerini, A. Vullo and P. Frasconi. DISULFIND: a Disulfide Bonding State and Cysteine Connectivity Prediction Server, Nucleic Acids Research, 34(Web Server issue):W177--W181, 2006.http://disulfind.dsi.unifi.it/
- ^ ExPASy Bioinformatics Resource Portal. Proteomics tool. http://expasy.org/proteomics
- ^ National Center for Biotechnology Information, U.S. National Library of Medicine website. GEO Profiles.https://www.ncbi.nlm.nih.gov/geoprofiles
- ^ a b Mahdessian H, Taxiarchis A, Popov S, Silveira A, Franco-Cereceda A, Hamsten A, Eriksson P, van't Hooft F (June 2014). "TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content". Proceedings of the National Academy of Sciences of the United States of America. 111 (24): 8913–8. Bibcode:2014PNAS..111.8913M. doi:10.1073/pnas.1323785111. PMC 4066487. PMID 24927523.
- ^ MINT program "MINT database". Archived from the original on 2006-05-06. Retrieved 2011-05-09.
- ^ IntAct program http://www.ebi.ac.uk/intact/
- ^ STRING program http://string.embl.de/
- ^ US patent office. Predictors of transplant rejection determined by peripheral blood gene-expression profiling. Publication number; US8053182 B2. Publication type; Grant. Application number; US 10/587,569. PCT number; PCT/US2005/002697. Publication date; Nov 8, 2011. Inventors; Thomas Cappola, Jonathan A. Epstein.
- ^ Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, Vogt TF, Hobbs HH, Cohen JC (April 2014). "Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease". Nature Genetics. 46 (4): 352–6. doi:10.1038/ng.2901. PMC 3969786. PMID 24531328.