Methanococcus maripaludis is a species of methanogenic archaea found in marine environments, predominantly salt marshes.[1] M. maripaludis is a non-pathogenic, gram-negative, weakly motile, non-spore-forming, and strictly anaerobic mesophile.[2] It is classified as a chemolithoautotroph.[3] This archaeon has a pleomorphic coccoid-rod shape of 1.2 by 1.6 μm, in average size, and has many unique metabolic processes that aid in survival.[2][4] M. maripaludis also has a sequenced genome consisting of around 1.7 Mbp with over 1,700 identified protein-coding genes.[5] In ideal conditions, M. maripaludis grows quickly and can double every two hours.[4]
| Methanococcus maripaludis | |
|---|---|
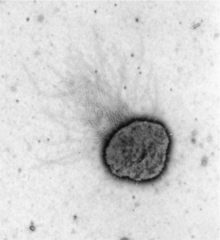
| |
| Electron micrograph showing prominent archaeal features and interior body of the microorganism. Courtesy of Dr. Ken F. Jarrell and Shin-Ichi Aizawa. Mag bar of 500nm. | |
| Scientific classification | |
| Domain: | |
| Kingdom: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | Methanococcus maripaludis Jones et al. 1984
|
Metabolism
editThe metabolic landscape of M. maripaludis consists of eight major subsystems which provide pathways for energy generation and cell growth. These subsystems include amino acid metabolism, glycolysis/glycogen metabolism, methanogenesis, nitrogen metabolism, non-oxidative pentose phosphate pathway (NOPPP), nucleotide metabolism, and the reductive citric acid (RTCA) cycle.[4]
Methanogenesis, the process of reducing carbon dioxide to methane, serves as the primary pathway for energy generation using coenzymes and a membrane-bound enzyme complex.[6] The methanogenesis pathway uses the same carbon source as the remaining seven subsystems for cell growth.[4] Additionally, the subsystems use two essential intermediates, acetyl CoA and pyruvate, to produce precursors critical for cell growth.[4]
Amino acid
editM. maripaludis uses carbon dioxide and acetate as substrates for amino acid biosynthesis.[4] Each of these substrates can produce Acetyl-CoA through various mechanisms.[4] Using carbon dioxide, M. maripaludis can generate Acetyl-CoA from methyl-THMPT, an intermediate of methanogenesis, and carbon monoxide, produced from the reduction of carbon dioxide.[4] Using acetate, Acetyl-CoA is synthesized from the AMP-forming acetate CoA ligase.[4] Acetyl-CoA then acts as a precursor to pyruvate, which promotes methanogenesis and alanine biosynthesis.[4] Pyruvate can be converted to L-alanine via alanine dehydrogenase, which is a reversible reaction. Once alanine is synthesized, it can be transported into the microbe via alanine permease.[4]
Glycolysis with formation of glycogen
editM. maripaludis has a modified Embden Meyerhof-Parnas (EMP) pathway, a glycolysis pathway. Dissimilarly to other organisms that reduce NAD to NADH in the EMP Pathway, M. maripaludis reduces ferrodoxins. Additionally, the protein kinases, responsible for transferring phosphate groups between compounds, uniquely rely on ADP rather than ATP.[4] Additionally, M. maripaludis is also capable of synthesizing glycogen.[4] Due to experimentally observed activities of enzymes involved in both the catabolic and anabolic directions of the EMP Pathway, the latter is utilized to a higher extent, resulting in the formation of glycogen stores.[4]
Methanogenesis
editIn M. maripaludis, the primary carbon source for methanogenesis is carbon dioxide, although alternatives such as formate are also used. Though all methanogens use certain key coenzymes, cofactors, and intermediates to produce methane, M. maripaludis undergoes the Wolfe cycle, which converts CO2 and hydrogen gas into methane and H2O.[7] 7 different hydrogenases are present in M. maripaludis that allow for the usage of H2 as an electron donor to reduce CO2.[4] Some strains and mutants of M. maripaludis have been shown to be capable of methanogenesis in the absence of hydrogen gas, though this is uncommon.[8]
Methanogenesis in M. maripaludis occurs in the following steps:
- Reduction of CO2 via methanofuran and reduced ferredoxins[9]
- Oxidation and subsequent reduction of the coenzyme F420 in the presence of H2[10][9]
- Transfer of a methyl group from methyl-THMPT to coenzyme M (HS-CoM), driving translocation of 2 Na+ across the membrane to strengthen the proton gradient[11]
- Demethylation of methyl-S-CoM to form methane and generate additional energy via the subsequent reduction of byproducts with H2[12]
Nitrogen
editM. maripaludis uses three sources of nitrogen: ammonia, alanine, and dinitrogen with ammonia.[4] Nitrogen assimilation occurs in the bacteria through ammonia when an inorganic nitrogen compound is converted to an organic nitrogen compound. In M. maripaludis, glutamine synthetase is used to make glutamine from glutamate and ammonia. The glutamine created then is sent to continue through protein synthesis.[4]
M. maripaludis uses alanine racemase and alanine permease for alanine uptake.[4] A racemase enzyme is used to convert the inversion of stereochemistry within the molecule while a permease is a protein that catalyzes the transport of a molecule across the membrane.[13]
Free N2 fixation is well established in M. maripaludis. M. maripaludis contains a multiprotein nitrogen complex containing an Fe protein and a MoFe.[4] The ferredoxin is reduced and reduces the oxidized Fe, stripping the Fe of its electrons in the presence of N2. The now reduced Fe protein is oxidized by ATP, reducing the MoFe protein.[4] The MoFe protein then reduces N2 to ammonia. This reductive step uses the electrons from the reduced ferredoxin which requires high amounts of energy. N2 fixation is unfavorable in M. maripaludis because of the high energy demand, so it is common for a cell to not activate this fixation pathway when ammonia and alanine are available.[4]
Pentose phosphate pathway
editThe pentose phosphate pathway is essential for M. maripaludis to make nucleotides and nucleic acids.[4] M. maripaludis contains high activities of non-oxidative enzymes, but has no oxidative enzyme activities.[4] Non-oxidative means that the enzymes do not have the ability to combine with oxygen and oxidize. The non-oxidative pentose phosphate pathway (NOPPP) is regulated and used through substrate availability. In M. maripaludis, ribulose-5-phosphate is converted to erythrose-4-phosphate and fructose-6-phosphate.[4] Four enzymes are used in this conversion: transketoloase, ribulose-phosphate 3-epimerase, ribose-5-phosphate isomerase, and translaldolase.[4]
Nucleotide metabolism
editNucleotide metabolism by M. maripaludis is well understood. For nucleic acid biosynthesis, the methanogen must produce pyrimidines, such as uridine triphosphate (UTP) and cytidine triphosphate (CTP), as well as purines such as guanine triphosphate (GTP) and adenosine triphosphate (ATP). To synthesize the pyrimidines, phosphoribosyl pyrophosphate (PRPP) combines with bicarbonate, L-glutamine, or orotate. This combination synthesizes uridine monophosphate, which can then be converted into uridine triphosphate (UTP). UMP also functions as a precursor to CTP.[4] To synthesize the purines, inosinic acid (IMP) is first made via a series of reactions, which include PRPP combining with glutamine to form 5-phosphoribosylamine. This reaction is catalyzed by PRPP synthetase. Once IMP is synthesized, it can be further converted into adenosine monophosphate (AMP) and guanine monophosphate (GMP). To synthesize AMP, IMP combines with adenylosuccinate. To synthesize GMP, IMP is converted into xanthine monophosphate (XMP) which can then be converted into GMP.[4]
Reductive Citric Acid (RTCA) Cycle
editThe tricarboxylic acid cycle serves as a central metabolic pathway in aerobic organisms. It plays an essential role in energy production and biosynthesis by generating electron carriers such as NADH and FAD.[14] This is performed by oxidizing acetyl-CoA, derived from various nutrients and complex carbon molecules, to CO2 and H2O.[4]
M. maripaludis, a strictly anaerobic mesophile, undergoes an incomplete Reductive Citric Acid (RTA) Cycle to reduce CO2 and H2O and synthesize complex carbon molecules.[4] Lacking several steps and essential enzymes, including phosphoenolpyruvate carboxykinase, citrate synthase, aconitate, and isocitrate dehydrogenase, hinders the completion of this cycle.[15][4] Pyruvate, produced from glycolysis/gluconeogenesis, is an initial metabolite in M. maripaludis for the Tricarboxylic Acid Cycle.
Cell structure
editThe irregular-shaped, weakly-motile coccus, Methanococcus maripaludis, has a diameter of 0.9-1.3 μm with a single, electron-dense S-layer lacking peptidoglycan. These characteristics assist in identifying its domain as Archaea.[4] Commonly found in methanogens, their cell walls lack murein and ether-linked membrane lipids, among other biochemical properties.[16] The S-layer is composed of glycoproteins that enclose the entire cell and help to protect the cell from direct interactions with the environment. More specifically, the S-layer provides archaeal cells a stabilization barrier that is resistant to environmental changes.[17] Additionally, M. maripaludis consists of two surface appendages assisting in motility: flagella and pili.[4]
Flagella and pili
editArchaeal flagella contain distinctive prokaryotic motility structures that are similar to bacterial type IV pili (T4P). They are constructed from proteins bearing class III signal peptides that are cleaved by specific signal peptidases. They also possess homologous genes, which encode an ATPase, and conserved membrane proteins for appendage assembly.[17] The flagellum of M. maripaludis is composed of three flagellin glycoproteins, which are all modified with an N-linked tetrasaccharide. This is critical for continued attachment to surfaces, cell-to-cell contact, and locomotion.[17] Both flagella and pili structures are used to attach to surfaces, allowing them the ability to remain in desirable environments.[17]
M. maripaludis encompasses a complete set of fla genes with three distinct flagellin genes, flaB1, flaB2 and flaB3, and the remaining eight genes including flaC-flaJ.[18] From the flagella locus, there are two major flagellin proteins required for flagella filaments, flaB1 and flaB2. Flagellin export also requires two specific proteins including flaH and flaI. The hook-like protein in M. maripaludis is strongly indicated by the minor flagellin protein, flaB3.[18] The flagella in numerous archaea undergo post-translational modifications, including glycosylation. Consequently, these flagella exhibit larger proteins than their expected gene sequence.[18]
Similar to the flagella, the proteins involved in the pilus assembly of M. maripaludis exhibit resemblance to bacterial Type IV pili due to the presence of an N-terminal signal peptide and an anticipated N-terminal hydrophobic α-helix.[19][20] The two pilin-like genes, MMP0236 (epdB) and MMP0237 (epdC), possess a short, atypical signal peptide ending in a conserved glycine. This is then succeeded by a hydrophobic segment, resulting in a distinct quaternary structure and pilus formation.[20]
Genetics
editMethanococcus maripaludis is one of four hydrogenotrophic methanogens, along with Methanocaldococcus jannaschii, Methanothermobacter thermautotrophicus, and Methanopyrus kandleri, to have its genome sequenced.[4] Of these three, Methanocaldococcus jannaschii is the closest living, known relative of M. maripaludis. M. maripaludis, like many other archaea, has one single circular chromosome.[4] According to the number of BlastP hits in the genome sequence, or similar protein sequences identified by the Basic Local Alignment Search Tool (BLAST), M. maripaludis is similar to most other methanogens.[4] However, M. maripaludis is missing common features, such as the ribulose bisphosphate carboxylase enzyme.[4]
Twenty one different strains of M. maripaludis have had their genomes sequenced, and each genome includes many copies of the chromosome in the singular cell, ranging from 5 to 55.[21] Of its 1,722 protein coding genes, 835 ORFs, or open reading frames, have unknown functions, and 129 ORFs are unique to M. maripaludis.[4] Some of these genes have been identified using in vivo transposon mutagenesis that may be essential for growth, making up approximately 30% of the genome.[22] The sequenced genome also revealed about 48 protein transporter systems, largely dominated by ABC transporters followed by iron transporters.[5]
M. maripaludis has been genetically altered to produce non-native, desired products, such as geraniol and polyhydroxybutyrate.[21] M. maripaludis can be used to sequence a variety of promoters and ribosome-binding sites using CRISPR/Cas9 technology.[23] Large deletions in the DNA can also be facilitated by a CRISPR/Cas9 system specifically designed for a strain named S0001.[21]
Environmental roles
editMethanogens play important roles in waste water treatment, carbon conversion, hydrogen production, and many other environmental processes.[4] In terms of waste water treatment, methanogens have been used to anaerobically degrade waste into methane utilizing a symbiotic relationship with syntrophic bacteria.[4] M. maripaludis, in addition to other methanogens, has the potential for generating fuels, such as methane and methanol, from CO2 emissions due to native CO2 uptake.[4] CO2 emissions are currently one of the leading sources of global warming. The ability of M. maripaludis to uptake CO2 from the environment in the presence of N2 allows for a potential route for conversion of CO2 emissions to a useful fuel like methane.[4] It is able to capture and convert CO2 from power and chemical plant emissions as well. Despite the many potential applications, the need for large amounts of hydrogen is an issue with using any methanogen for biomethane production.[4]
References
edit- ^ Franklin MJ, Wiebe WJ, Whitman WB (May 1988). "Populations of methanogenic bacteria in a georgia salt marsh". Applied and Environmental Microbiology. 54 (5): 1151–1157. Bibcode:1988ApEnM..54.1151F. doi:10.1128/aem.54.5.1151-1157.1988. PMC 202619. PMID 16347628.
- ^ a b Jones WJ, Paynter MJ, Gupta R (1983-08-01). "Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment". Archives of Microbiology. 135 (2): 91–97. Bibcode:1983ArMic.135...91J. doi:10.1007/BF00408015. ISSN 1432-072X.
- ^ Müller AL, Gu W, Patsalo V, Deutzmann JS, Williamson JR, Spormann AM (April 2021). "An alternative resource allocation strategy in the chemolithoautotrophic archaeon Methanococcus maripaludis". Proceedings of the National Academy of Sciences of the United States of America. 118 (16). Bibcode:2021PNAS..11825854M. doi:10.1073/pnas.2025854118. PMC 8072206. PMID 33879571.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap Goyal N, Zhou Z, Karimi IA (June 2016). "Metabolic processes of Methanococcus maripaludis and potential applications". Microbial Cell Factories. 15 (1): 107. doi:10.1186/s12934-016-0500-0. PMC 4902934. PMID 27286964.
- ^ a b Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, et al. (October 2004). "Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis". Journal of Bacteriology. 186 (20): 6956–6969. doi:10.1128/JB.186.20.6956-6969.2004. PMC 522202. PMID 15466049.
- ^ Liu Y, Whitman WB (March 2008) [26 March 2008]. "Metabolic, Phylogenetic, and Ecological Diversity of the Methanogenic Archaea". Annals of the New York Academy of Sciences. 1125 (1): 171–189. Bibcode:2008NYASA1125..171L. doi:10.1196/annals.1419.019. ISSN 0077-8923. PMID 18378594.
- ^ Escalante-Semerena JC, Rinehart KL, Wolfe RS (August 1984). "Tetrahydromethanopterin, a carbon carrier in methanogenesis". The Journal of Biological Chemistry. 259 (15): 9447–9455. doi:10.1016/s0021-9258(17)42721-9. PMID 6547718.
- ^ Lohner ST, Deutzmann JS, Logan BE, Leigh J, Spormann AM (August 2014). "Hydrogenase-independent uptake and metabolism of electrons by the archaeon Methanococcus maripaludis". The ISME Journal. 8 (8): 1673–1681. Bibcode:2014ISMEJ...8.1673L. doi:10.1038/ismej.2014.82. PMC 4817615. PMID 24844759.
- ^ a b Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R (August 2008). "Methanogenic archaea: ecologically relevant differences in energy conservation". Nature Reviews. Microbiology. 6 (8): 579–591. doi:10.1038/nrmicro1931. PMID 18587410. S2CID 32698014.
- ^ Mukhopadhyay B, Stoddard SF, Wolfe RS (February 1998). "Purification, regulation, and molecular and biochemical characterization of pyruvate carboxylase from Methanobacterium thermoautotrophicum strain deltaH". The Journal of Biological Chemistry. 273 (9): 5155–5166. doi:10.1074/jbc.273.9.5155. PMID 9478969.
- ^ Kengen SW, Daas PJ, Duits EF, Keltjens JT, van der Drift C, Vogels GD (February 1992). "Isolation of a 5-hydroxybenzimidazolyl cobamide-containing enzyme involved in the methyltetrahydromethanopterin: coenzyme M methyltransferase reaction in Methanobacterium thermoautotrophicum". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1118 (3): 249–260. doi:10.1016/0167-4838(92)90282-i. PMID 1737047.
- ^ Kaster AK, Moll J, Parey K, Thauer RK (February 2011). "Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea". Proceedings of the National Academy of Sciences of the United States of America. 108 (7): 2981–2986. Bibcode:2011PNAS..108.2981K. doi:10.1073/pnas.1016761108. PMC 3041090. PMID 21262829.
- ^ "Definition of PERMEASE". www.merriam-webster.com. Retrieved 2024-03-17.
- ^ Ladapo J, Whitman WB (August 1990). "Method for isolation of auxotrophs in the methanogenic archaebacteria: role of the acetyl-CoA pathway of autotrophic CO2 fixation in Methanococcus maripaludis". Proceedings of the National Academy of Sciences of the United States of America. 87 (15): 5598–5602. Bibcode:1990PNAS...87.5598L. doi:10.1073/pnas.87.15.5598. ISSN 0027-8424. PMC 54374. PMID 11607093.
- ^ Shieh JS, Whitman WB (November 1987). "Pathway of acetate assimilation in autotrophic and heterotrophic methanococci". Journal of Bacteriology. 169 (11): 5327–5329. doi:10.1128/jb.169.11.5327-5329.1987. ISSN 0021-9193. PMC 213948. PMID 3667534.
- ^ Jarrell KF, Koval SF (1989). "Ultrastructure and biochemistry of Methanococcus voltae". Critical Reviews in Microbiology. 17 (1): 53–87. doi:10.3109/10408418909105722. ISSN 1040-841X. PMID 2669831.
- ^ a b c d Jarrell KF, Stark M, Nair DB, Chong JP (June 2011). "Flagella and pili are both necessary for efficient attachment of Methanococcus maripaludis to surfaces". FEMS Microbiology Letters. 319 (1): 44–50. doi:10.1111/j.1574-6968.2011.02264.x. PMID 21410509. S2CID 36895781.
- ^ a b c d Chaban B, Ng SY, Kanbe M, Saltzman I, Nimmo G, Aizawa SI, et al. (November 2007). "Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis". Molecular Microbiology. 66 (3): 596–609. doi:10.1111/j.1365-2958.2007.05913.x. ISSN 0950-382X. PMID 17887963.
- ^ Szabó Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJ, Pohlschröder M (February 2007). "Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases". Journal of Bacteriology. 189 (3): 772–778. doi:10.1128/JB.01547-06. ISSN 0021-9193. PMC 1797317. PMID 17114255.
- ^ a b Wang YA, Yu X, Ng SY, Jarrell KF, Egelman EH (2008-08-29). "The structure of an archaeal pilus". Journal of Molecular Biology. 381 (2): 456–466. doi:10.1016/j.jmb.2008.06.017. ISSN 1089-8638. PMC 2570433. PMID 18602118.
- ^ a b c Li J, Akinyemi TS, Shao N, Chen C, Dong X, Liu Y, et al. (2023). "Genetic and metabolic engineering of Methanococcus spp". Current Research in Biotechnology. 5: 100115. doi:10.1016/j.crbiot.2022.11.002. ISSN 2590-2628.
- ^ Sarmiento F, Mrázek J, Whitman WB (2013-03-19). "Genome-scale analysis of gene function in the hydrogenotrophic methanogenic archaeon Methanococcus maripaludis". Proceedings of the National Academy of Sciences. 110 (12): 4726–4731. Bibcode:2013PNAS..110.4726S. doi:10.1073/pnas.1220225110. ISSN 0027-8424. PMC 3607031. PMID 23487778.
- ^ Xu Q, Du Q, Gao J, Chen L, Dong X, Li J (2023-07-24). "A robust genetic toolbox for fine-tuning gene expression in the CO2-Fixing methanogenic archaeon Methanococcus maripaludis". Metabolic Engineering. 79: 130–145. doi:10.1016/j.ymben.2023.07.007. ISSN 1096-7176. PMID 37495072.
Further reading
edit- Haydock AK, Porat I, Whitman WB, Leigh JA (September 2004). "Continuous culture of Methanococcus maripaludis under defined nutrient conditions". FEMS Microbiology Letters. 238 (1): 85–91. doi:10.1016/j.femsle.2004.07.021. PMID 15336407.