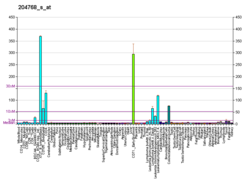
Flap endonuclease 1 is an enzyme that in humans is encoded by the FEN1 gene.[5][6]
Function
editThe protein encoded by this gene removes 5' overhanging "flaps" (or short sections of single stranded DNA that "hang off" because their nucleotide bases are prevented from binding to their complementary base pair—despite any base pairing downstream) in DNA repair and processes the 5' ends of Okazaki fragments in lagging strand DNA synthesis. Direct physical interaction between this protein and AP endonuclease 1 during long-patch base excision repair provides coordinated loading of the proteins onto the substrate, thus passing the substrate from one enzyme to another. The protein is a member of the XPG/RAD2 endonuclease family and is one of ten proteins essential for cell-free DNA replication. DNA secondary structure can inhibit flap processing at certain trinucleotide repeats in a length-dependent manner by concealing the 5' end of the flap that is necessary for both binding and cleavage by the protein encoded by this gene. Therefore, secondary structure can deter the protective function of this protein, leading to site-specific trinucleotide expansions.[6]
Interactions
editFlap structure-specific endonuclease 1 has been shown to interact with:
Over expression of FEN1 in cancers
editFEN1 is over-expressed in the majority of cancers of the breast,[17] prostate,[18] stomach,[19][20] neuroblastomas,[21] pancreatic,[22] and lung.[23]
FEN1 is an essential enzyme in an inaccurate pathway for repair of double-strand breaks in DNA called microhomology-dependent alternative end joining or microhomology-mediated end joining (MMEJ).[24] MMEJ always involves at least a small deletion, so that it is a mutagenic pathway.[25] Several other pathways can also repair double-strand breaks in DNA, including the less inaccurate pathway of non-homologous end joining (NHEJ) and accurate pathways using homologous recombinational repair (HRR).[26] Various factors determine which pathway will be used for repair of double strand breaks in DNA.[25] When FEN1 is over-expressed (this occurs when its promoter is hypomethylated[17]) the highly inaccurate MMEJ pathway may be favored, causing a higher rate of mutation and increased risk of cancer.
Cancers are very often deficient in expression of one or more DNA repair genes, but over-expression of a DNA repair gene is unusual in cancer. For instance, at least 36 DNA repair enzymes, when mutationally defective in germ line cells, cause increased risk of cancer (hereditary cancer syndromes).[citation needed] Similarly, at least 12 DNA repair genes have frequently been found to be epigenetically repressed in one or more cancers.[citation needed] (See also Epigenetically reduced DNA repair and cancer.) Ordinarily, deficient expression of a DNA repair enzyme results in increased un-repaired DNA damages which, through replication errors (translesion synthesis), lead to mutations and cancer. However, FEN1 mediated MMEJ repair is highly inaccurate, so in this case, over-expression, rather than under-expression, leads to cancer.
Therapeutic target for human cancer
editTherapeutic targets for cancers with BRCA1 or BRCA2 defects were identified by analysis of synthetic lethal relationships using Saccharomyces cerevisiae, human cell lines and mice as model systems.[27] Inhibition of the FEN1 repair protein with small molecule inhibitors was observed to preferentially kill cancer cell lines that were already deficient in expression of BRCA1 and BRCA2 proteins. Cancers that often have defective expression of BRCA1 or BRCA2 include breast cancer and ovarian cancer. Such cancers are deficient in the DNA repair process of homologous recombination (HR). FEN1 protein is essential for the alternate DNA repair pathway, microhomology-mediated end joining (MMEJ). Thus, FEN1 inhibition of the MMEJ repair pathway of cancer cells, that are already defective in the HR repair pathway, causes a second repair pathway to be deficient leading to synthetic lethality.
References
edit- ^ a b c GRCh38: Ensembl release 89: ENSG00000168496 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000024742 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Hiraoka LR, Harrington JJ, Gerhard DS, Lieber MR, Hsieh CL (January 1995). "Sequence of human FEN-1, a structure-specific endonuclease, and chromosomal localization of the gene (FEN1) in mouse and human". Genomics. 25 (1): 220–225. doi:10.1016/0888-7543(95)80129-A. PMID 7774922.
- ^ a b "Entrez Gene: FEN1 flap structure-specific endonuclease 1".
- ^ a b Dianova II, Bohr VA, Dianov GL (October 2001). "Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair". Biochemistry. 40 (42): 12639–12644. doi:10.1021/bi011117i. PMID 11601988.
- ^ a b Sharma S, Sommers JA, Wu L, Bohr VA, Hickson ID, Brosh RM (March 2004). "Stimulation of flap endonuclease-1 by the Bloom's syndrome protein". The Journal of Biological Chemistry. 279 (11): 9847–9856. doi:10.1074/jbc.M309898200. PMID 14688284.
- ^ a b c Henneke G, Koundrioukoff S, Hübscher U (July 2003). "Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation". Oncogene. 22 (28): 4301–4313. doi:10.1038/sj.onc.1206606. PMID 12853968. S2CID 5787920.
- ^ a b Hasan S, Stucki M, Hassa PO, Imhof R, Gehrig P, Hunziker P, et al. (June 2001). "Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300". Molecular Cell. 7 (6): 1221–1231. doi:10.1016/s1097-2765(01)00272-6. PMID 11430825.
- ^ Chai Q, Zheng L, Zhou M, Turchi JJ, Shen B (December 2003). "Interaction and stimulation of human FEN-1 nuclease activities by heterogeneous nuclear ribonucleoprotein A1 in alpha-segment processing during Okazaki fragment maturation". Biochemistry. 42 (51): 15045–15052. doi:10.1021/bi035364t. PMID 14690413.
- ^ Jónsson ZO, Hindges R, Hübscher U (April 1998). "Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen". The EMBO Journal. 17 (8): 2412–2425. doi:10.1093/emboj/17.8.2412. PMC 1170584. PMID 9545252.
- ^ Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS (September 1997). "The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21". The Journal of Biological Chemistry. 272 (39): 24522–24529. doi:10.1074/jbc.272.39.24522. PMID 9305916.
- ^ Chen U, Chen S, Saha P, Dutta A (October 1996). "p21Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex". Proceedings of the National Academy of Sciences of the United States of America. 93 (21): 11597–11602. Bibcode:1996PNAS...9311597C. doi:10.1073/pnas.93.21.11597. PMC 38103. PMID 8876181.
- ^ Yu P, Huang B, Shen M, Lau C, Chan E, Michel J, et al. (January 2001). "p15(PAF), a novel PCNA associated factor with increased expression in tumor tissues". Oncogene. 20 (4): 484–489. doi:10.1038/sj.onc.1204113. PMID 11313979. S2CID 39144360.
- ^ Brosh RM, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, et al. (October 2001). "Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity". The EMBO Journal. 20 (20): 5791–5801. doi:10.1093/emboj/20.20.5791. PMC 125684. PMID 11598021.
- ^ a b Singh P, Yang M, Dai H, Yu D, Huang Q, Tan W, et al. (November 2008). "Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers". Molecular Cancer Research. 6 (11): 1710–1717. doi:10.1158/1541-7786.MCR-08-0269. PMC 2948671. PMID 19010819.
- ^ Lam JS, Seligson DB, Yu H, Li A, Eeva M, Pantuck AJ, et al. (August 2006). "Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score". BJU International. 98 (2): 445–451. doi:10.1111/j.1464-410X.2006.06224.x. PMID 16879693. S2CID 22165252.
- ^ Kim JM, Sohn HY, Yoon SY, Oh JH, Yang JO, Kim JH, et al. (January 2005). "Identification of gastric cancer-related genes using a cDNA microarray containing novel expressed sequence tags expressed in gastric cancer cells". Clinical Cancer Research. 11 (2 Pt 1): 473–482. doi:10.1158/1078-0432.473.11.2. PMID 15701830.
- ^ Wang K, Xie C, Chen D (May 2014). "Flap endonuclease 1 is a promising candidate biomarker in gastric cancer and is involved in cell proliferation and apoptosis". International Journal of Molecular Medicine. 33 (5): 1268–1274. doi:10.3892/ijmm.2014.1682. PMID 24590400.
- ^ Krause A, Combaret V, Iacono I, Lacroix B, Compagnon C, Bergeron C, et al. (July 2005). "Genome-wide analysis of gene expression in neuroblastomas detected by mass screening" (PDF). Cancer Letters. 225 (1): 111–120. doi:10.1016/j.canlet.2004.10.035. PMID 15922863. S2CID 44644467.
- ^ Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, et al. (April 2003). "Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays". The American Journal of Pathology. 162 (4): 1151–1162. doi:10.1016/S0002-9440(10)63911-9. PMC 1851213. PMID 12651607.
- ^ Nikolova T, Christmann M, Kaina B (July 2009). "FEN1 is overexpressed in testis, lung and brain tumors". Anticancer Research. 29 (7): 2453–2459. PMID 19596913.
- ^ Sharma S, Javadekar SM, Pandey M, Srivastava M, Kumari R, Raghavan SC (March 2015). "Homology and enzymatic requirements of microhomology-dependent alternative end joining". Cell Death & Disease. 6 (3): e1697. doi:10.1038/cddis.2015.58. PMC 4385936. PMID 25789972.
- ^ a b Liang L, Deng L, Chen Y, Li GC, Shao C, Tischfield JA (September 2005). "Modulation of DNA end joining by nuclear proteins". The Journal of Biological Chemistry. 280 (36): 31442–31449. doi:10.1074/jbc.M503776200. PMID 16012167.
- ^ Ottaviani D, LeCain M, Sheer D (March 2014). "The role of microhomology in genomic structural variation". Trends in Genetics. 30 (3): 85–94. doi:10.1016/j.tig.2014.01.001. PMID 24503142.
- ^ Guo E, Ishii Y, Mueller J, Srivatsan A, Gahman T, Putnam CD, et al. (August 2020). "FEN1 endonuclease as a therapeutic target for human cancers with defects in homologous recombination". Proceedings of the National Academy of Sciences of the United States of America. 117 (32): 19415–19424. Bibcode:2020PNAS..11719415G. doi:10.1073/pnas.2009237117. PMC 7431096. PMID 32719125.
Further reading
edit- Finger LD, Blanchard MS, Theimer CA, Sengerová B, Singh P, Chavez V, et al. (August 2009). "The 3'-flap pocket of human flap endonuclease 1 is critical for substrate binding and catalysis". The Journal of Biological Chemistry. 284 (33): 22184–22194. doi:10.1074/jbc.M109.015065. PMC 2755943. PMID 19525235.
- Kemeny MM, Alava G, Oliver JM (November 1992). "Improving responses in hepatomas with circadian-patterned hepatic artery infusions of recombinant interleukin-2". Journal of Immunotherapy. 12 (4): 219–223. doi:10.1097/00002371-199211000-00001. PMID 1477073.
- Li X, Li J, Harrington J, Lieber MR, Burgers PM (September 1995). "Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen". The Journal of Biological Chemistry. 270 (38): 22109–22112. doi:10.1074/jbc.270.38.22109. PMID 7673186.
- Robins P, Pappin DJ, Wood RD, Lindahl T (November 1994). "Structural and functional homology between mammalian DNase IV and the 5'-nuclease domain of Escherichia coli DNA polymerase I". The Journal of Biological Chemistry. 269 (46): 28535–28538. doi:10.1016/S0021-9258(19)61935-6. PMID 7961795.
- Murray JM, Tavassoli M, al-Harithy R, Sheldrick KS, Lehmann AR, Carr AM, Watts FZ (July 1994). "Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage". Molecular and Cellular Biology. 14 (7): 4878–4888. doi:10.1128/MCB.14.7.4878. PMC 358860. PMID 8007985.
- Harrington JJ, Lieber MR (March 1994). "The characterization of a mammalian DNA structure-specific endonuclease". The EMBO Journal. 13 (5): 1235–1246. doi:10.1002/j.1460-2075.1994.tb06373.x. PMC 394933. PMID 8131753.
- Shen B, Nolan JP, Sklar LA, Park MS (April 1996). "Essential amino acids for substrate binding and catalysis of human flap endonuclease 1". The Journal of Biological Chemistry. 271 (16): 9173–9176. doi:10.1074/jbc.271.16.9173. PMID 8621570.
- Chen U, Chen S, Saha P, Dutta A (October 1996). "p21Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex". Proceedings of the National Academy of Sciences of the United States of America. 93 (21): 11597–11602. Bibcode:1996PNAS...9311597C. doi:10.1073/pnas.93.21.11597. PMC 38103. PMID 8876181.
- Warbrick E, Lane DP, Glover DM, Cox LS (May 1997). "Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: a potential mechanism to co-ordinate DNA replication and repair". Oncogene. 14 (19): 2313–2321. doi:10.1038/sj.onc.1201072. PMID 9178907. S2CID 29525059.
- Klungland A, Lindahl T (June 1997). "Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1)". The EMBO Journal. 16 (11): 3341–3348. doi:10.1093/emboj/16.11.3341. PMC 1169950. PMID 9214649.
- Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS (September 1997). "The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21". The Journal of Biological Chemistry. 272 (39): 24522–24529. doi:10.1074/jbc.272.39.24522. PMID 9305916.
- Stöhr H, Marquardt A, Rivera A, Cooper PR, Nowak NJ, Shows TB, et al. (January 1998). "A gene map of the Best's vitelliform macular dystrophy region in chromosome 11q12-q13.1". Genome Research. 8 (1): 48–56. doi:10.1101/gr.8.1.48. PMC 310689. PMID 9445487.
- Jónsson ZO, Hindges R, Hübscher U (April 1998). "Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen". The EMBO Journal. 17 (8): 2412–2425. doi:10.1093/emboj/17.8.2412. PMC 1170584. PMID 9545252.
- Warbrick E, Heatherington W, Lane DP, Glover DM (September 1998). "PCNA binding proteins in Drosophila melanogaster : the analysis of a conserved PCNA binding domain". Nucleic Acids Research. 26 (17): 3925–3932. doi:10.1093/nar/26.17.3925. PMC 147798. PMID 9705499.
- Hosfield DJ, Mol CD, Shen B, Tainer JA (October 1998). "Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity". Cell. 95 (1): 135–146. doi:10.1016/S0092-8674(00)81789-4. PMID 9778254. S2CID 8283941.
- Dianov GL, Jensen BR, Kenny MK, Bohr VA (August 1999). "Replication protein A stimulates proliferating cell nuclear antigen-dependent repair of abasic sites in DNA by human cell extracts". Biochemistry. 38 (34): 11021–11025. doi:10.1021/bi9908890. PMID 10460157.
- Greene AL, Snipe JR, Gordenin DA, Resnick MA (November 1999). "Functional analysis of human FEN1 in Saccharomyces cerevisiae and its role in genome stability" (PDF). Human Molecular Genetics. 8 (12): 2263–2273. doi:10.1093/hmg/8.12.2263. PMID 10545607.
- Matsumoto Y, Kim K, Hurwitz J, Gary R, Levin DS, Tomkinson AE, Park MS (November 1999). "Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins". The Journal of Biological Chemistry. 274 (47): 33703–33708. doi:10.1074/jbc.274.47.33703. PMID 10559261.
- Spiro C, Pelletier R, Rolfsmeier ML, Dixon MJ, Lahue RS, Gupta G, et al. (December 1999). "Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats". Molecular Cell. 4 (6): 1079–1085. doi:10.1016/S1097-2765(00)80236-1. PMID 10635332.
- Hasan S, Stucki M, Hassa PO, Imhof R, Gehrig P, Hunziker P, et al. (June 2001). "Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300". Molecular Cell. 7 (6): 1221–1231. doi:10.1016/S1097-2765(01)00272-6. PMID 11430825.
- Caldwell RB, Braselmann H, Schoetz U, Heuer S, Scherthan H, Zitzelsberger H (July 2016). "Positive Cofactor 4 (PC4) is critical for DNA repair pathway re-routing in DT40 cells". Scientific Reports. 6: 28890. Bibcode:2016NatSR...628890C. doi:10.1038/srep28890. PMC 4931448. PMID 27374870.