Darobactin is an experimental antibiotic compound that may be effective against Gram-negative bacteria. If it can be developed into a human-compatible form it would be the first to come from an animal microbiome.[1]
| |
| Names | |
|---|---|
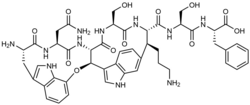
| IUPAC name
(2S)-2-[[(2S)-2-[[(5R,17S,20S,26S,29S,30S)-17-amino-20-(2-amino-2-oxoethyl)-30-(3-aminopropyl)-26-(hydroxymethyl)-18,21,24,27-tetraoxo-6-oxa-2,13,19,22,25,28-hexazahexacyclo[29.3.1.04,34.05,23.07,12.011,15]pentatriaconta-1(34),3,7,9,11,14,31(35),32-octaene-29-carbonyl]amino]-3-hydroxypropanoyl]amino]-3-phenylpropanoic acid
| |
| Identifiers | |
3D model (JSmol)
|
|
PubChem CID
|
|
| |
| |
| Properties | |
| C47H55N11O12 | |
| Molar mass | 966.022 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
History
editThe compound was discovered in Photorhabdus bacteria in 2019, living in the digestive systems of entomopathogenic nematodes.[2] Researchers identified the nematode as a possible host because they feed on insects by targeting their larvae and releasing bacteria that then confront pathogens similar to those found in humans.[1]
In experiments, it cured mice of infections with Escherichia coli and Klebsiella pneumoniae, both members of the Enterobacteriaceae, without toxic side effects.[1]
Gram-negative bacteria
editGram-negative bacteria have a characteristic architecture for the cell envelope, with an inner membrane, an outer membrane, and a periplasmic space in between. In this arrangement, the peptidoglycan layer is relatively thin and does not retain the crystal violet stain used in the Gram staining method of bacterial classification. Antibiotic resistance has become widespread in bacterial pathogens, and in Gram-negative bacteria such as the Enterobacteriaceae, much of this comes from acquired genes. The resistance genes encode proteins that export or inactivate β-lactam antibiotics, aminoglycosides, tetracycline, chloramphenicol, fosfomycin, etc. Plasmids carrying these genes readily move between strains or between species. Consequently, resistance to the currently available panel of approved antibiotics is an increasingly worrisome problem.[1] The most recent class of antibiotics effective against these bacteria emerged in the 1960s.[2]
Mode of action
editDarobactin inhibits BamA and disrupts the proper formation of the Gram-negative cell envelope.[2] BamA is a central component of the BamABCDE complex, which inserts proteins from the periplasm into the outer membrane. BamA also aids in the folding of outer membrane-bound proteins. Thus, darobactin prevents the proper formation of the outer membrane of bacteria, leading to cell death.[1] Because only Gram-negative bacteria have BamA and outer membrane beta-barrel proteins, only they are susceptible to darobactin.
Biosynthesis
editDarobactin is a RiPP, that is, a ribosomally synthesized and post-translationally modified peptide. Its production and export is encoded by a typically silent five gene operon that showed minimal production under laboratory culture conditions. Mature darobactin consists of a seven amino acids core peptide derived from a longer precursor, with unusual Trp-1 to Trp-3 and Trp-3 to Lys-5 (or Arg-5, in variant forms) crosslinks.[2] The key enzyme for maturation from the precursor to the mature form is the radical SAM/SPASM enzyme DarE.
References
edit- ^ a b c d e Nield, David (24 November 2019). "A Tiny Worm Has Been Found Carrying New Antibiotic That Could Help Us Fight Superbugs". ScienceAlert. Archived from the original on 2019-11-25. Retrieved 2019-11-26.
- ^ a b c d Imai, Yu; Meyer, Kirsten J.; Iinishi, Akira; Favre-Godal, Quentin; Green, Robert; Manuse, Sylvie; Caboni, Mariaelena; Mori, Miho; Niles, Samantha; Ghiglieri, Meghan; Honrao, Chandrashekhar (2019-11-20). "A new antibiotic selectively kills Gram-negative pathogens". Nature. 576 (7787): 459–464. Bibcode:2019Natur.576..459I. doi:10.1038/s41586-019-1791-1. ISSN 1476-4687. PMC 7188312. PMID 31747680.