DNA (cytosine-5)-methyltransferase 3A (DNMT3A) is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. The enzyme is encoded in humans by the DNMT3A gene.[5][6]
This enzyme is responsible for de novo DNA methylation. Such function is to be distinguished from maintenance DNA methylation which ensures the fidelity of replication of inherited epigenetic patterns. DNMT3A forms part of the family of DNA methyltransferase enzymes, which consists of the protagonists DNMT1, DNMT3A and DNMT3B.[5][6]
While de novo DNA methylation modifies the information passed on by the parent to the progeny, it enables key epigenetic modifications essential for processes such as cellular differentiation and embryonic development, transcriptional regulation, heterochromatin formation, X-inactivation, imprinting and genome stability.[7]
DNMT3a is the gene most commonly found mutated in clonal hematopoiesis, a common aging-related phenomenon in which hematopoietic stem cells (HSCs) or other early blood cell progenitors contribute to the formation of a genetically distinct subpopulation of blood cells.[8][9][10]
Gene
editDNMT3A is a 130 kDa protein encoded by 23 exons found on chromosome 2p23 in humans.[11] There exists a 98% homology between human and murine homologues.[6] DNMT3A is widely expressed among mammals.[12]
There are two main protein isoforms, DNMT3A1 and DNMT3A2 with molecular weights of about 130 kDa and 100 kDa, respectively. The DNMT3A2 protein, which lacks the N-terminal region of DNMT3A1, is encoded by a transcript initiated from a downstream promoter.[13] These isoforms exist in different cell types.[14] When originally established,[13] DNMT3A2 was found to be highly expressed in testis, ovary, spleen, and thymus. It was more recently shown to be inducibly expressed in brain hippocampus[15] and needed in the hippocampus when establishing memory.[16] DNMT3A2 is also upregulated in the nucleus accumbens shell in response to cocaine.[17]
Protein structure
editDNMT3A consists of three major protein domains: the Pro-Trp-Trp-Pro (PWWP) domain, the ATRX-DNMT3-DNMT3L (ADD) domain and the catalytic methyltransferase domain.
The structures of DNMT3A1 and DNMT3A2 have analogies with the structure of DNMT3B1 and also with the two accessory proteins DNMT3B3 and DNMT3L (see Figure of simplified domains of DNMT3A isoforms). The two accessory proteins stimulate de novo methylation by each of their interactions with the three isoforms that have a functional catalytic domain. In general, all DNMTs require accessory proteins for their biological function.[18]
The PWWP motif is within an about 100 amino acid domain that has one area with a significant amount of basic residues (lysines and arginines), giving a positively charged surface that can bind to DNA. A separate region of the PWWP domain can bind to histone methyl-lysines through a hydrophobic pocket that includes the PWWP motif itself.[19][20]
The ADD domain of DNMT3A is composed of an N-terminal GATA-like zinc finger, a PHD finger and a C-terminal alpha helix, which, together, are arranged into a single globular fold. This domain can act as a reader that specifically binds to histone H3 that is unmethylated at lysine 4 (H3K4me0).[21] The ADD domain serves as an inhibitor of the methyltransferase domain until DNMT3A binds to the unmodified lysine 4 of histone 3 (H3K4me0) for its de novo methylating activity.[14] DNMT3A thus seems to have an inbuilt control mechanism targeting DNA for methylation only at histones that are unmethylated at histone 3 with the lysine at the 4th position from the amino end being un-methylated.
The catalytic domain (the methyltransferase domain) is highly conserved, even among prokaryotes.[22]
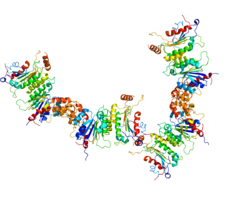
The three DNA methyltransferases (DNMT3A1, DNMT3A2 and DNMT3B) catalyze reactions placing a methyl group onto a cytosine, usually at a CpG site in DNA.[23] The accompanying Figure shows a methyltransferase complex containing DNMT3A2. These enzymes, to be effective, must act in conjunction with an accessory protein (e.g. DNMT3B3, DNMT3L, or others).[24][25][26] Two accessory proteins (which have no catalytic activity), complexed to two DNMTs with a catalytic domain, occur as a heterotetramer (see Figure). These heterotetramers occur in the order: accessory protein-catalytic protein-catalytic protein-accessory protein. The particular complex shown in the Figure illustrates the heterotetramer formed by catalytic protein DNMT3A2 and accessory protein DNMT3B3. One accessory protein of the complex binds to an acidic patch on the nucleosome core (see top 3B3 in Figure). The connection of one accessory protein to the nucleosome orients the heterotetramer. The orientation places the first catalytic DNMT (closest to the accessory protein connected to the nucleosome) in an intermediate position (not close to the linker DNA). The second catalytic DNMT (lower 3A2 in Figure) is placed at the linker DNA. Methylations can take place within this linker DNA (as shown in the Figure) but not on any DNA wrapped around the nucleosome core.
As shown by Manzo et al.,[27] there are both specific individual binding sites for the three catalytic DNMTs (3A1, 3A2 and 3B3) as well as overlapping binding sites of these enzymes. There are 28 million CpG sites in the human genome.[28] Many of these CpGs are located within CpG islands (regions of DNA) of relatively high density of CpG sites.[28] Of these regions, there are 3,970 regions exclusively enriched for DNMT3A1, 3,838 regions for DNMT3A2 and 3,432 regions for DNMT3B, and there are sites that are shared between the de novo DNMT proteins.[27] In addition, whether the DNA methyltransferase (DNMT3A1, DNMT3A2 or DNMT3B) acts on an available CpG site depends on the sequence flanking the CpG site within the linker DNA.[26]
Function
editDNMT1 is responsible for maintenance DNA methylation while DNMT3A and DNMT3B carry out both maintenance – correcting the errors of DNMT1 – and de novo DNA methylation. After DNMT1 knockout in human cancer cells, these cells were found to retain their inherited methylation pattern,[29] which suggests maintenance activity by the expressed DNMT3s. DNMT3s show equal affinity for unmethylated and hemimethylated DNA substrates[29] while DNMT1 has a 10-40 fold preference for hemimethylated DNA.[30][31] The DNMT3s can bind to both forms and hence potentially do both maintenance and de novo modifications.
De novo methylation is the main recognized activity of DNMT3A, which is essential for processes such as those mentioned in the introductory paragraphs. Genetic imprinting prevents parthenogenesis in mammals,[32] and hence forces sexual reproduction and its multiple consequences on genetics and phylogenesis. DNMT3A is essential for genetic imprinting.[33]
Research on long-term memory storage in humans indicates that memory is maintained by DNA methylation,[34] Rats in which a new, strong long-term memory is induced due to contextual fear conditioning have reduced expression of about 1,000 genes and increased expression of about 500 genes in the hippocampus region of the brain. These changes occur 24 hours after training. At this point, there is modified expression of 9.17% of the rat hippocampal genome. Reduced expression of genes is associated with de novo methylations of the genes.[35]
Animal studies
editIn mice, this gene has shown reduced expression in ageing animals causes cognitive long-term memory decline.[15]
In Dnmt3a-/- mice, many genes associated with HSC self-renewal increase in expression and some fail to be appropriately repressed during differentiation.[36] This suggests abrogation of differentiation in hematopoietic stem cells (HSCs) and an increase in self-renewal cell-division instead. Indeed, it was found that differentiation was partially rescued if Dnmt3a-/- HSCs experienced an additional Ctnb1 knockdown – Ctnb1 codes for β-catenin, which participates in self-renewal cell division.[14]
Clinical relevance
editThis gene is frequently mutated in cancer, being one of 127 frequently mutated genes identified in the Cancer Genome Atlas project[37] DNMT3A mutations were most commonly seen in acute myeloid leukaemia (AML) where they occurred in just over 25% of cases sequenced. These mutations most often occur at position R882 in the protein and this mutation may cause loss of function.[38] DNMT3A mutations are associated with poor overall survival, suggesting that they have an important common effect on the potential of AML cells to cause lethal disease.[39] It has also been found that DNMT3A-mutated cell lines exhibit transcriptome instability, in that they have much more erroneous RNA splicing as compared to their isogenic wildtype counterparts.[40] Mutations in this gene are also associated with Tatton-Brown–Rahman syndrome, an overgrowth disorder.
Interactions
editDNMT3A has been shown to interact with:
References
edit- ^ a b c GRCh38: Ensembl release 89: ENSG00000119772 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000020661 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b Okano M, Xie S, Li E (July 1998). "Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases". Nature Genetics. 19 (3): 219–220. doi:10.1038/890. PMID 9662389. S2CID 256263.
- ^ a b c Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, et al. (August 1999). "Cloning, expression and chromosome locations of the human DNMT3 gene family". Gene. 236 (1): 87–95. doi:10.1016/S0378-1119(99)00252-8. PMID 10433969.
- ^ Jia Y, Li P, Fang L, Zhu H, Xu L, Cheng H, et al. (2016-04-12). "Negative regulation of DNMT3A de novo DNA methylation by frequently overexpressed UHRF family proteins as a mechanism for widespread DNA hypomethylation in cancer". Cell Discovery. 2: 16007. doi:10.1038/celldisc.2016.7. PMC 4849474. PMID 27462454.
- ^ Jan M, Ebert BL, Jaiswal S (January 2017). "Clonal hematopoiesis". Seminars in Hematology. 54 (1): 43–50. doi:10.1053/j.seminhematol.2016.10.002. PMC 8045769. PMID 28088988.
- ^ Sperling AS, Gibson CJ, Ebert BL (January 2017). "The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia". Nature Reviews. Cancer. 17 (1): 5–19. doi:10.1038/nrc.2016.112. PMC 5470392. PMID 27834397.
- ^ Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL (July 2015). "Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes". Blood. 126 (1): 9–16. doi:10.1182/blood-2015-03-631747. PMC 4624443. PMID 25931582.
- ^ Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA (June 1999). "The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors". Nucleic Acids Research. 27 (11): 2291–2298. doi:10.1093/nar/27.11.2291. PMC 148793. PMID 10325416.
- ^ Del Castillo Falconi VM, Torres-Arciga K, Matus-Ortega G, Díaz-Chávez J, Herrera LA (August 2022). "DNA Methyltransferases: From Evolution to Clinical Applications". International Journal of Molecular Sciences. 23 (16): 8994. doi:10.3390/ijms23168994. PMC 9409253. PMID 36012258.
- ^ a b Chen T, Ueda Y, Xie S, Li E (October 2002). "A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation". The Journal of Biological Chemistry. 277 (41): 38746–38754. doi:10.1074/jbc.M205312200. PMID 12138111.
- ^ a b c Yang L, Rau R, Goodell MA (March 2015). "DNMT3A in haematological malignancies". Nature Reviews. Cancer. 15 (3): 152–165. doi:10.1038/nrc3895. PMC 5814392. PMID 25693834.
- ^ a b Oliveira AM, Hemstedt TJ, Bading H (July 2012). "Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities". Nature Neuroscience. 15 (8): 1111–1113. doi:10.1038/nn.3151. PMID 22751036. S2CID 10590208.
- ^ Oliveira AM, Hemstedt TJ, Freitag HE, Bading H (August 2016). "Dnmt3a2: a hub for enhancing cognitive functions". Molecular Psychiatry. 21 (8): 1130–1136. doi:10.1038/mp.2015.175. PMID 26598069. S2CID 25308306.
- ^ Cannella N, Oliveira AM, Hemstedt T, Lissek T, Buechler E, Bading H, Spanagel R (August 2018). "Dnmt3a2 in the Nucleus Accumbens Shell Is Required for Reinstatement of Cocaine Seeking". The Journal of Neuroscience. 38 (34): 7516–7528. doi:10.1523/JNEUROSCI.0600-18.2018. PMC 6596133. PMID 30030395.
- ^ Gujar H, Weisenberger DJ, Liang G (February 2019). "The Roles of Human DNA Methyltransferases and Their Isoforms in Shaping the Epigenome". Genes. 10 (2): 172. doi:10.3390/genes10020172. PMC 6409524. PMID 30813436.
- ^ van Nuland R, van Schaik FM, Simonis M, van Heesch S, Cuppen E, Boelens R, et al. (May 2013). "Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain". Epigenetics & Chromatin. 6 (1): 12. doi:10.1186/1756-8935-6-12. PMC 3663649. PMID 23656834.
- ^ Rona GB, Eleutherio EC, Pinheiro AS (March 2016). "PWWP domains and their modes of sensing DNA and histone methylated lysines". Biophysical Reviews. 8 (1): 63–74. doi:10.1007/s12551-015-0190-6. PMC 5425739. PMID 28510146.
- ^ Ren W, Gao L, Song J (December 2018). "Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation". Genes. 9 (12): 620. doi:10.3390/genes9120620. PMC 6316889. PMID 30544982.
- ^ Xu F, Mao C, Ding Y, Rui C, Wu L, Shi A, et al. (2010-01-01). "Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs". Current Medicinal Chemistry. 17 (33): 4052–4071. doi:10.2174/092986710793205372. PMC 3003592. PMID 20939822.
- ^ Zeng Y, Ren R, Kaur G, Hardikar S, Ying Z, Babcock L, Gupta E, Zhang X, Chen T, Cheng X (November 2020). "The inactive Dnmt3b3 isoform preferentially enhances Dnmt3b-mediated DNA methylation". Genes Dev. 34 (21–22): 1546–1558. doi:10.1101/gad.341925.120. PMC 7608744. PMID 33004415.
- ^ Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X (September 2007). "Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation". Nature. 449 (7159): 248–51. Bibcode:2007Natur.449..248J. doi:10.1038/nature06146. PMC 2712830. PMID 17713477.
- ^ Xu TH, Liu M, Zhou XE, Liang G, Zhao G, Xu HE, Melcher K, Jones PA (October 2020). "Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B". Nature. 586 (7827): 151–155. Bibcode:2020Natur.586..151X. doi:10.1038/s41586-020-2747-1. PMC 7540737. PMID 32968275.
- ^ a b Gao L, Emperle M, Guo Y, Grimm SA, Ren W, Adam S, Uryu H, Zhang ZM, Chen D, Yin J, Dukatz M, Anteneh H, Jurkowska RZ, Lu J, Wang Y, Bashtrykov P, Wade PA, Wang GG, Jeltsch A, Song J (July 2020). "Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms". Nat Commun. 11 (1): 3355. Bibcode:2020NatCo..11.3355G. doi:10.1038/s41467-020-17109-4. PMC 7335073. PMID 32620778.
- ^ a b Manzo M, Wirz J, Ambrosi C, Villaseñor R, Roschitzki B, Baubec T (December 2017). "Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands". EMBO J. 36 (23): 3421–3434. doi:10.15252/embj.201797038. PMC 5709737. PMID 29074627.
- ^ a b Lövkvist C, Dodd IB, Sneppen K, Haerter JO (June 2016). "DNA methylation in human epigenomes depends on local topology of CpG sites". Nucleic Acids Res. 44 (11): 5123–32. doi:10.1093/nar/gkw124. PMC 4914085. PMID 26932361.
- ^ a b Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, et al. (April 2000). "CpG methylation is maintained in human cancer cells lacking DNMT1". Nature. 404 (6781): 1003–1007. Bibcode:2000Natur.404.1003R. doi:10.1038/35010000. PMID 10801130. S2CID 4425037.
- ^ Pradhan S, Bacolla A, Wells RD, Roberts RJ (November 1999). "Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation". The Journal of Biological Chemistry. 274 (46): 33002–33010. doi:10.1074/jbc.274.46.33002. PMID 10551868.
- ^ Pradhan S, Talbot D, Sha M, Benner J, Hornstra L, Li E, et al. (November 1997). "Baculovirus-mediated expression and characterization of the full-length murine DNA methyltransferase". Nucleic Acids Research. 25 (22): 4666–4673. doi:10.1093/nar/25.22.4666. PMC 147102. PMID 9358180.
- ^ Reik W, Walter J (January 2001). "Genomic imprinting: parental influence on the genome". Nature Reviews. Genetics. 2 (1): 21–32. doi:10.1038/35047554. PMID 11253064. S2CID 12050251.
- ^ Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H (June 2004). "Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting". Nature. 429 (6994): 900–903. Bibcode:2004Natur.429..900K. doi:10.1038/nature02633. PMID 15215868. S2CID 4344982.
- ^ Miller CA, Sweatt JD (March 2007). "Covalent modification of DNA regulates memory formation". Neuron. 53 (6): 857–869. doi:10.1016/j.neuron.2007.02.022. PMID 17359920.
- ^ Duke CG, Kennedy AJ, Gavin CF, Day JJ, Sweatt JD (July 2017). "Experience-dependent epigenomic reorganization in the hippocampus". Learn Mem. 24 (7): 278–288. doi:10.1101/lm.045112.117. PMC 5473107. PMID 28620075.
- ^ Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. (December 2011). "Dnmt3a is essential for hematopoietic stem cell differentiation". Nature Genetics. 44 (1): 23–31. doi:10.1038/ng.1009. PMC 3637952. PMID 22138693.
- ^ Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. (October 2013). "Mutational landscape and significance across 12 major cancer types". Nature. 502 (7471): 333–339. Bibcode:2013Natur.502..333K. doi:10.1038/nature12634. PMC 3927368. PMID 24132290.
- ^ Shih AH, Abdel-Wahab O, Patel JP, Levine RL (September 2012). "The role of mutations in epigenetic regulators in myeloid malignancies". Nature Reviews. Cancer. 12 (9): 599–612. doi:10.1038/nrc3343. PMID 22898539. S2CID 20214444.
- ^ Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. (December 2010). "DNMT3A mutations in acute myeloid leukemia". The New England Journal of Medicine. 363 (25): 2424–2433. doi:10.1056/NEJMoa1005143. PMC 3201818. PMID 21067377.
- ^ Banaszak LG, Giudice V, Zhao X, Wu Z, Gao S, Hosokawa K, et al. (March 2018). "Abnormal RNA splicing and genomic instability after induction of DNMT3A mutations by CRISPR/Cas9 gene editing". Blood Cells, Molecules & Diseases. 69: 10–22. doi:10.1016/j.bcmd.2017.12.002. PMC 6728079. PMID 29324392.
- ^ a b Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S (August 2002). "Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases". The EMBO Journal. 21 (15): 4183–4195. doi:10.1093/emboj/cdf401. PMC 126147. PMID 12145218.
- ^ a b c d Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD (2004). "Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription". Nucleic Acids Research. 32 (2): 598–610. doi:10.1093/nar/gkh195. PMC 373322. PMID 14752048.
- ^ Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. (July 2003). "Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin". Current Biology. 13 (14): 1192–1200. Bibcode:2003CBio...13.1192L. doi:10.1016/s0960-9822(03)00432-9. PMID 12867029. S2CID 2320997.
- ^ a b Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T (May 2001). "Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription". The EMBO Journal. 20 (10): 2536–2544. doi:10.1093/emboj/20.10.2536. PMC 125250. PMID 11350943.
- ^ Brenner C, Deplus R, Didelot C, Loriot A, Viré E, De Smet C, et al. (January 2005). "Myc represses transcription through recruitment of DNA methyltransferase corepressor". The EMBO Journal. 24 (2): 336–346. doi:10.1038/sj.emboj.7600509. PMC 545804. PMID 15616584.
- ^ Fuks F, Hurd PJ, Deplus R, Kouzarides T (May 2003). "The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase". Nucleic Acids Research. 31 (9): 2305–2312. doi:10.1093/nar/gkg332. PMC 154218. PMID 12711675.
Further reading
edit- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, et al. (September 1995). "Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence". Nature. 377 (6547 Suppl): 3–174. PMID 7566098.
- Bonaldo MF, Lennon G, Soares MB (September 1996). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Research. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA (June 1999). "The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors". Nucleic Acids Research. 27 (11): 2291–2298. doi:10.1093/nar/27.11.2291. PMC 148793. PMID 10325416.
- Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T (May 2001). "Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription". The EMBO Journal. 20 (10): 2536–2544. doi:10.1093/emboj/20.10.2536. PMC 125250. PMID 11350943.
- Vaute O, Nicolas E, Vandel L, Trouche D (January 2002). "Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases". Nucleic Acids Research. 30 (2): 475–481. doi:10.1093/nar/30.2.475. PMC 99834. PMID 11788710.
- Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, et al. (February 2002). "Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor". Science. 295 (5557): 1079–1082. Bibcode:2002Sci...295.1079D. doi:10.1126/science.1065173. hdl:11576/2506625. PMID 11834837. S2CID 29532358.
- Hata K, Okano M, Lei H, Li E (April 2002). "Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice". Development. 129 (8): 1983–1993. doi:10.1242/dev.129.8.1983. PMID 11934864.
- Chen T, Ueda Y, Xie S, Li E (October 2002). "A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation". The Journal of Biological Chemistry. 277 (41): 38746–38754. doi:10.1074/jbc.M205312200. PMID 12138111.
- Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S (August 2002). "Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases". The EMBO Journal. 21 (15): 4183–4195. doi:10.1093/emboj/cdf401. PMC 126147. PMID 12145218.
- Yanagisawa Y, Ito E, Yuasa Y, Maruyama K (September 2002). "The human DNA methyltransferases DNMT3A and DNMT3B have two types of promoters with different CpG contents". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1577 (3): 457–465. doi:10.1016/S0167-4781(02)00482-7. PMID 12359337.
- Fatemi M, Hermann A, Gowher H, Jeltsch A (October 2002). "Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA". European Journal of Biochemistry. 269 (20): 4981–4984. doi:10.1046/j.1432-1033.2002.03198.x. PMID 12383256.
- Weisenberger DJ, Velicescu M, Preciado-Lopez MA, Gonzales FA, Tsai YC, Liang G, Jones PA (September 2002). "Identification and characterization of alternatively spliced variants of DNA methyltransferase 3a in mammalian cells". Gene. 298 (1): 91–99. doi:10.1016/S0378-1119(02)00976-9. PMID 12406579.
- Chedin F, Lieber MR, Hsieh CL (December 2002). "The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a". Proceedings of the National Academy of Sciences of the United States of America. 99 (26): 16916–16921. Bibcode:2002PNAS...9916916C. doi:10.1073/pnas.262443999. PMC 139244. PMID 12481029.
- Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR (January 2003). "DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells". Nature Genetics. 33 (1): 61–65. doi:10.1038/ng1068. PMID 12496760. S2CID 561490.
- Fuks F, Hurd PJ, Deplus R, Kouzarides T (May 2003). "The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase". Nucleic Acids Research. 31 (9): 2305–2312. doi:10.1093/nar/gkg332. PMC 154218. PMID 12711675.
- Yakushiji T, Uzawa K, Shibahara T, Noma H, Tanzawa H (June 2003). "Over-expression of DNA methyltransferases and CDKN2A gene methylation status in squamous cell carcinoma of the oral cavity". International Journal of Oncology. 22 (6): 1201–1207. doi:10.3892/ijo.22.6.1201. PMID 12738984.
- Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD (2004). "Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription". Nucleic Acids Research. 32 (2): 598–610. doi:10.1093/nar/gkh195. PMC 373322. PMID 14752048.