The fluid mosaic model explains various characteristics regarding the structure of functional cell membranes. According to this biological model, there is a lipid bilayer (two molecules thick layer consisting primarily of amphipathic phospholipids) in which protein molecules are embedded. The phospholipid bilayer gives fluidity and elasticity to the membrane. Small amounts of carbohydrates are also found in the cell membrane. The biological model, which was devised by Seymour Jonathan Singer and Garth L. Nicolson in 1972,[1] describes the cell membrane as a two-dimensional liquid where embedded proteins are generally randomly distributed. For example, it is stated that "A prediction of the fluid mosaic model is that the two-dimensional long-range distribution of any integral protein in the plane of the membrane is essentially random."[1]
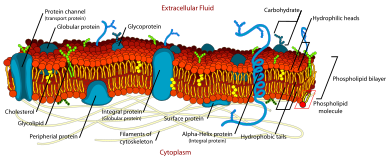
Chemical makeup
edit| Components | Location | Functions |
|---|---|---|
| Phospholipid | The main fabric of plasma membrane | It provides selective permeability to the cell membrane.
The hydrophilic phosphate side is outwards and hydrophobic inwards. |
| Carbohydrates | Attached to proteins on outside membrane layers | It helps in cell-to-cell recognition. |
| Cholesterol | Between phospholipids and phospholipid bilayers | It helps the plasma membrane to retain its fluidity. |
| Proteins | Embedded within or on the surface of phospholipid layers | These form channels to allow the movement of molecules. |
Experimental evidence
editThe fluid property of functional biological membranes had been determined through labeling experiments, x-ray diffraction, and calorimetry. These studies showed that integral membrane proteins diffuse at rates affected by the viscosity of the lipid bilayer in which they were embedded, and demonstrated that the molecules within the cell membrane are dynamic rather than static.[1]
Previous models of biological membranes included the Robertson Unit Membrane Model and the Davson-Danielli Tri-Layer model.[2] These models had proteins present as sheets neighboring a lipid layer, rather than incorporated into the phospholipid bilayer. Other models described repeating, regular units of protein and lipid. These models were not well supported by microscopy and thermodynamic data, and did not accommodate evidence for dynamic membrane properties.[2]
An important experiment that provided evidence supporting fluid and dynamic biological was performed by Frye and Edidin. They used Sendai virus to force human and mouse cells to fuse and form a heterokaryon. Using antibody staining, they were able to show that the mouse and human proteins remained segregated to separate halves of the heterokaryon a short time after cell fusion. However, the proteins eventually diffused and over time the border between the two halves was lost. Lowering the temperature slowed the rate of this diffusion by causing the membrane phospholipids to transition from a fluid to a gel phase.[3] Singer and Nicolson rationalized the results of these experiments using their fluid mosaic model.[1]
The fluid mosaic model explains changes in structure and behavior of cell membranes under different temperatures, as well as the association of membrane proteins with the membranes. While Singer and Nicolson had substantial evidence drawn from multiple subfields to support their model, recent advances in fluorescence microscopy and structural biology have validated the fluid mosaic nature of cell membranes.
Subsequent developments
editMembrane asymmetry
editAdditionally, the two leaflets of biological membranes are asymmetric and divided into subdomains composed of specific proteins or lipids, allowing spatial segregation of biological processes associated with membranes. Cholesterol and cholesterol-interacting proteins can concentrate into lipid rafts and constrain cell signaling processes to only these rafts.[4] Another form of asymmetry was shown by the work of Mouritsen and Bloom in 1984, where they proposed a Mattress Model of lipid-protein interactions to address the biophysical evidence that the membrane can range in thickness and hydrophobicity of proteins.[5]
Non-bilayer membranes
editThe existence of non-bilayer lipid formations with important biological functions was confirmed subsequent to publication of the fluid mosaic model. These membrane structures may be useful when the cell needs to propagate a non bilayer form, which occurs during cell division and the formation of a gap junction.[6]
Membrane curvature
editThe membrane bilayer is not always flat. Local curvature of the membrane can be caused by the asymmetry and non-bilayer organization of lipids as discussed above. More dramatic and functional curvature is achieved through BAR domains, which bind to phosphatidylinositol on the membrane surface, assisting in vesicle formation, organelle formation and cell division.[7] Curvature development is in constant flux and contributes to the dynamic nature of biological membranes.[8]
Lipid movement within the membrane
editDuring the 1970s, it was acknowledged that individual lipid molecules undergo free lateral diffusion within each of the layers of the lipid membrane.[9] Diffusion occurs at a high speed, with an average lipid molecule diffusing ~2μm, approximately the length of a large bacterial cell, in about 1 second.[9] It has also been observed that individual lipid molecules rotate rapidly around their own axis.[9] Moreover, phospholipid molecules can, although they seldom do, migrate from one side of the lipid bilayer to the other (a process known as flip-flop). However, flip-flop movement is enhanced by flippase enzymes.[10] The processes described above influence the disordered nature of lipid molecules and interacting proteins in the lipid membranes, with consequences to membrane fluidity, signaling, trafficking and function.
Restrictions to lateral diffusion
editThere are restrictions to the lateral mobility of the lipid and protein components in the fluid membrane imposed by zonation. Early attempts to explain the assembly of membrane zones include the formation of lipid rafts and “cytoskeletal fences”, corrals wherein lipid and membrane proteins can diffuse freely, but that they can seldom leave.[2] These ideas remain controversial, and alternative explanations are available such as the proteolipid code.[11]
Lipid rafts
editLipid rafts are membrane nanometric platforms with a particular lipid and protein composition that laterally diffuse, navigating on the liquid bilipid layer. Sphingolipids and cholesterol are important building blocks of the lipid rafts.[12]
Protein complexes
editCell membrane proteins and glycoproteins do not exist as single elements of the lipid membrane, as first proposed by Singer and Nicolson in 1972. Rather, they occur as diffusing complexes within the membrane.[2] The assembly of single molecules into these macromolecular complexes has important functional consequences for the cell; such as ion and metabolite transport, signaling, cell adhesion, and migration.[2]
Cytoskeletal fences (corrals) and binding to the extracellular matrix
editSome proteins embedded in the bilipid layer interact with the extracellular matrix outside the cell, cytoskeleton filaments inside the cell, and septin ring-like structures. These interactions have a strong influence on shape and structure, as well as on compartmentalization. Moreover, they impose physical constraints that restrict the free lateral diffusion of proteins and at least some lipids within the bilipid layer.[2]
When integral proteins of the lipid bilayer are tethered to the extracellular matrix, they are unable to diffuse freely. Proteins with a long intracellular domain may collide with a fence formed by cytoskeleton filaments.[13] Both processes restrict the diffusion of proteins and lipids directly involved, as well as of other interacting components of the cell membranes.
Septin ring-like structures (in green) can pinch cell membranes and split them into subdomains.
Septins are a family of GTP-binding proteins highly conserved among eukaryotes. Prokaryotes have similar proteins called paraseptins. They form compartmentalizing ring-like structures strongly associated with the cell membranes. Septins are involved in the formation of structures such as, cilia and flagella, dendritic spines, and yeast buds.[14]
Historical timeline
edit- 1895 – Ernest Overton hypothesized that cell membranes are made out of lipids.[15]
- 1925 – Evert Gorter and François Grendel found that red blood cell membranes are formed by a fatty layer two molecules thick, i.e. they described the bilipid nature of the cell membrane.[16]
- 1935 – Hugh Davson and James Danielli proposed that lipid membranes are layers composed by proteins and lipids with pore-like structures that allow specific permeability for certain molecules. Then, they suggested a model for the cell membrane, consisting of a lipid layer surrounded by protein layers at both sides of it.[17]
- 1957 – J. David Robertson, based on electron microscopy studies, establishes the "Unit Membrane Hypothesis". This, states that all membranes in the cell, i.e. plasma and organelle membranes, have the same structure: a bilayer of phospholipids with monolayers of proteins at both sides of it.[18]
- 1972 – SJ Singer and GL Nicolson proposed the fluid mosaic model as an explanation for the data and latest evidence regarding the structure and thermodynamics of cell membranes.[1]
- 1997 – K Simons and E Ikonen proposed the lipid raft theory as an initial explanation of membrane zonation.[19]
- 2024 – TA Kervin and M Overduin proposed the proteolipid code to fully explain membrane zonation as the lipid raft theory became increasingly controversial.[20][21]
Notes and references
edit- ^ a b c d e Singer SJ, Nicolson GL (February 1972). "The fluid mosaic model of the structure of cell membranes". Science. 175 (4023): 720–731. Bibcode:1972Sci...175..720S. doi:10.1126/science.175.4023.720. PMID 4333397. S2CID 83851531.
- ^ a b c d e f Nicolson GL (June 2014). "The Fluid-Mosaic Model of Membrane Structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1838 (6): 1451–1466. doi:10.1016/j.bbamem.2013.10.019. PMID 24189436.
- ^ Frye LD, Edidin M (September 1970). "The rapid intermixing of cell surface antigens after formation of mouse-human heterokaryons". Journal of Cell Science. 7 (2): 319–335. doi:10.1242/jcs.7.2.319. PMID 4098863.
- ^ Silvius JR (December 2005). "Partitioning of membrane molecules between raft and non-raft domains: insights from model-membrane studies". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1746 (3): 193–202. doi:10.1016/j.bbamcr.2005.09.003. PMID 16271405.
- ^ Mouritsen OG, Bloom M (August 1984). "Mattress model of lipid-protein interactions in membranes". Biophysical Journal. 46 (2): 141–153. Bibcode:1984BpJ....46..141M. doi:10.1016/S0006-3495(84)84007-2. PMC 1435039. PMID 6478029.
- ^ van den Brink-van der Laan E, Killian JA, de Kruijff B (November 2004). "Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1666 (1–2): 275–288. doi:10.1016/j.bbamem.2004.06.010. PMID 15519321.
- ^ Frost A, Unger VM, De Camilli P (April 2009). "The BAR domain superfamily: membrane-molding macromolecules". Cell. 137 (2): 191–196. doi:10.1016/j.cell.2009.04.010. PMC 4832598. PMID 19379681.
- ^ Rodríguez-García R, Arriaga LR, Mell M, Moleiro LH, López-Montero I, Monroy F (March 2009). "Bimodal spectrum for the curvature fluctuations of bilayer vesicles: pure bending plus hybrid curvature-dilation modes". Physical Review Letters. 102 (12): 128101. Bibcode:2009PhRvL.102l8101R. doi:10.1103/PhysRevLett.102.128101. PMID 19392326.
- ^ a b c Alberts B, Johnson A, Lewis J, et al. (2008). Molecular Biology of the Cell (5th ed.). New York: Garland Science. pp. 621–622. ISBN 978-0-8153-4105-5.
- ^ Hankins HM, Baldridge RD, Xu P, Graham TR (January 2015). "Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution". Traffic. 16 (1): 35–47. doi:10.1111/tra.12233. PMC 4275391. PMID 25284293.
- ^ Christie, William (Bill) W. "Sphingolipids, Membrane Rafts and Caveolae - sphingomyelin and cholesterol - structure, occurrence, biochemistry and function". lipidmaps.org. Retrieved 14 September 2024.
- ^ Lingwood D, Simons K (January 2010). "Lipid rafts as a membrane-organizing principle". Science. 327 (5961): 46–50. Bibcode:2010Sci...327...46L. doi:10.1126/science.1174621. PMID 20044567. S2CID 35095032.
- ^ Vereb G, Szöllosi J, Matkó J, Nagy P, Farkas T, Vigh L, et al. (July 2003). "Dynamic, yet structured: The cell membrane three decades after the Singer-Nicolson model". Proceedings of the National Academy of Sciences of the United States of America. 100 (14): 8053–8058. Bibcode:2003PNAS..100.8053V. doi:10.1073/pnas.1332550100. PMC 166180. PMID 12832616.
- ^ Saarikangas J, Barral Y (October 2011). "The emerging functions of septins in metazoans". EMBO Reports. 12 (11): 1118–1126. doi:10.1038/embor.2011.193. PMC 3207108. PMID 21997296.
- ^ Overton E (1895). "Uberdie osmotischen Eigenshafter der Lebenden Pflanzen und tierzelle". VJSCHR Naturf Ges Zurich. 40: 159–201.
- ^ Gorter E, Grendel F (March 1925). "On Bimolecular Layers of Lipoids on the Chromocytes of the Blood". The Journal of Experimental Medicine. 41 (4): 439–443. doi:10.1084/jem.41.4.439. PMC 2130960. PMID 19868999.
- ^ Danielli J, Davson H (1935). "A contribution to the theory of permeability of thin films". Journal of Cellular and Comparative Physiology. 5 (4): 495–508. doi:10.1002/jcp.1030050409.
- ^ Heuser JE (1995). "In Memory of J. David Robertson" (PDF). Newsletter of the American Society of Cell Biology. Archived from the original (PDF) on 2018-10-08. Retrieved 2014-12-05.
- ^ Simons, Kai; Ikonen, Elina (June 1997). "Functional rafts in cell membranes". Nature. 387 (6633): 569–572. Bibcode:1997Natur.387..569S. doi:10.1038/42408. PMID 9177342.
- ^ Kervin, Troy A.; Overduin, Michael (27 February 2024). "Membranes are functionalized by a proteolipid code". BMC Biology. 22 (1): 46. doi:10.1186/s12915-024-01849-6. PMC 10898092. PMID 38414038.
- ^ Leslie, Mitch (25 November 2011). "Do Lipid Rafts Exist?". Science. 334 (6059): 1046–1047. Bibcode:2011Sci...334.1046L. doi:10.1126/science.334.6059.1046-b. PMID 22116853.