This article may be too technical for most readers to understand. (January 2024) |
The homing endonucleases are a collection of endonucleases encoded either as freestanding genes within introns, as fusions with host proteins, or as self-splicing inteins. They catalyze the hydrolysis of genomic DNA within the cells that synthesize them, but do so at very few, or even singular, locations. Repair of the hydrolyzed DNA by the host cell frequently results in the gene encoding the homing endonuclease having been copied into the cleavage site, hence the term 'homing' to describe the movement of these genes. Homing endonucleases can thereby transmit their genes horizontally within a host population, increasing their allele frequency at greater than Mendelian rates.
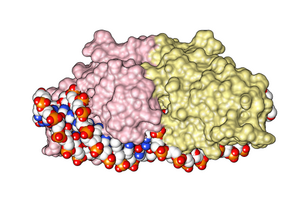
Origin and mechanism
editAlthough the origin and function of homing endonucleases is still being researched, the most established hypothesis considers them as selfish genetic elements,[1] similar to transposons, because they facilitate the perpetuation of the genetic elements that encode them independent of providing a functional attribute to the host organism.
Homing endonuclease recognition sequences are long enough to occur randomly only with a very low probability (approximately once every 7×109 bp),[2] and are normally found in one or very few instances per genome. Generally, owing to the homing mechanism, the gene encoding the endonuclease (the HEG, "homing endonuclease gene") is located within the recognition sequence which the enzyme cuts, thus interrupting the homing endonuclease recognition sequence and limiting DNA cutting only to sites that do not (yet) carry the HEG.
Prior to transmission, one allele carries the gene (HEG+) while the other does not (HEG−), and is therefore susceptible to being cut by the enzyme. Once the enzyme is synthesized, it breaks the chromosome in the HEG− allele, initiating a response from the cellular DNA repair system. The damage is repaired using recombination, taking the pattern of the opposite, undamaged DNA allele, HEG+, that contains the gene for the endonuclease. Thus, the gene is copied to the allele that initially did not have it and it is propagated through successive generations.[3] This process is called "homing".[3]
Nomenclature
editHoming endonucleases are always indicated with a prefix that identifies their genomic origin, followed by a hyphen: "I-" for homing endonucleases encoded within an intron, "PI-" (for "protein insert") for those encoded within an intein. Some authors have proposed using the prefix "F-" ("freestanding") for viral enzymes and other natural enzymes not encoded by introns nor inteins,[4] and "H-" ("hybrid") for enzymes synthesized in a laboratory.[5] Next, a three-letter name is derived from the binominal name of the organism, taking one uppercase letter from the genus name and two lowercase letters from the specific name. (Some mixing is usually done for hybrid enzymes.) Finally, a Roman numeral distinguishes different enzymes found in the same organism:
- PI-TliII (P30317) is the second-identified enzyme encoded by an intein found in the archaea Thermococcus litoralis.[6][7][8]
- H-DreI (PDB: 1MOW) is the first synthetic homing endonuclease, created in a laboratory from the enzymes I-DmoI (P21505) and I-CreI (P05725), taken respectively from Desulfurococcus mobilis and Chlamydomonas reinhardtii.[5][9]
Comparison to restriction enzymes
editHoming endonucleases differ from Type II restriction enzymes in the several respects:[4]
- Whereas Type II restriction enzymes bind short, usually symmetric, recognition sequences of 4 to 8 bp, homing endonucleases bind very long and in many cases asymmetric recognition sequences spanning 12 to 40 bp.
- Homing endonucleases are generally more tolerant of substitutions in the recognition sequence. Minor variations in the recognition sequence usually decrease the activity of homing endonucleases, but often do not completely abolish it as often occurs with restriction enzymes.[10][11]
- Homing endonucleases share structural motifs that suggest there are four families, whereas it has not been possible to determine simply recognisable and distinguishable families of Type II restriction enzymes.
- Homing endonucleases act as monomers or homodimers, and often require associated proteins to regulate their activity[12] or form ribonucleoprotein complexes, wherein RNA is an integral component of the catalytic apparatus.[13] Type II restriction enzymes can also function alone, as monomers or homodimers,[14] or with additional protein subunits,[15] but the accessory subunits differ from those of the homing endonucleases. Thus, they can require restriction, modification, and specificity subunits for their action.[15]
- Finally, homing endonucleases have a broader phylogenetic distribution, occurring in all three biological domains—the archaea, bacteria and eukarya. Type II restriction enzymes occur only in archaea, bacteria and certain viruses.[16][17][18] Homing endonucleases are also expressed in all three compartments of the eukaryotic cell: nuclei, mitochondria and chloroplasts. Open reading frames encoding homing endonucleases have been found in introns, inteins, and in freestanding form between genes, whereas genes encoding Type II restriction enzyme genes have been found only in freestanding form, almost always in close association with genes encoding cognate DNA modifying enzymes.[19] Thus, while the Type II restriction enzymes and homing endonucleases share the function of cleaving double-stranded DNA, they appear to have evolved independently.
Structural families
edit
| ||||||||||||||||
| LAGLIDADG endonuclease | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | LAGLIDADG_1 | ||||||||
| Pfam | PF00961 | ||||||||
| Pfam clan | CL0324 | ||||||||
| InterPro | IPR001982 | ||||||||
| CATH | 1af5 | ||||||||
| SCOP2 | 1af5 / SCOPe / SUPFAM | ||||||||
| |||||||||
| See clan entry for related Pfam families. | |||||||||
| GIY-YIG endonuclease, catalytic | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | GIY-YIG | ||||||||
| Pfam | PF01541 | ||||||||
| InterPro | IPR000305 | ||||||||
| PROSITE | PS50164 | ||||||||
| CATH | 1mk0 | ||||||||
| SCOP2 | 1mk0 / SCOPe / SUPFAM | ||||||||
| |||||||||
Currently there are six known structural families. Their conserved structural motifs are:[4]
- LAGLIDADG: Every polypeptide has 1 or 2 LAGLIDADG motifs. The sequence LAGLIDADG is a conserved sequence of amino acids where each letter is a code that identifies a specific residue. This sequence is directly involved in the DNA cutting process. Those enzymes that have only one motif work as homodimers, creating a saddle that interacts with the major groove of each DNA half-site. The LAGLIDADG motifs contribute amino acid residues to both the protein-protein interface between protein domains or subunits, and to the enzyme's active sites. Enzymes that possess two motifs in a single protein chain act as monomers, creating the saddle in a similar way. The first structures to be determined of homing endonucleases (of PI-SceI and I-CreI, both reported in 1997) were both from the LAGLIDADG structural family.,[20][21] The following year, the first structure of a homing endonuclease (I-CreI) bound to its DNA target site was also reported.[9]
- GIY-YIG: These have only one GIY-YIG motif, in the N-terminal region, that interacts with the DNA in the cutting site. The prototypic enzyme of this family is I-TevI which acts as a monomer. Separate structural studies have been reported of the DNA-binding and catalytic domains of I-TevI, the former bound to its DNA target and the latter in the absence of DNA.[22][23]
- His-Cys box (Pfam PF05551): These enzymes possess a region of 30 amino acids that includes 5 conserved residues: two histidines and three cysteins. They co-ordinate the metal cation needed for catalysis. I-PpoI is the best characterized enzyme of this family and acts as a homodimer. Its structure was reported in 1998.[24] It is possibly related to the H-N-H family, as they share common features.[25]
- H-N-H: (Pfam CL0263): These have a consensus sequence of approximately 30 amino acids. It includes two pairs of conserved histidines and one asparagine that create a zinc finger domain. I-HmuI (P34081) is the best characterized enzyme of this family, and acts as a monomer. Its structure was reported in 2004 (PDB: 1U3E).[26]
- PD-(D/E)xK (Pfam CL0236): These enzymes contain a canonical nuclease catalytic domain typically found in type II restriction endonucleases. The best characterized enzyme in this family, I-Ssp6803I (Q57253), acts as a tetramer. Its structure was reported in 2007 (PDB: 2OST).[27] The overall fold is conserved in many endonuclease families, all of which belong to the PD-(D/E)xK superfamily.[28]
- Vsr-like/EDxHD (DUF559, InterPro: IPR007569): These enzymes were discovered in the Global Ocean Sampling Metagenomic Database and first described in 2009. The term 'Vsr-like' refers to the presence of a C-terminal nuclease domain that displays recognizable homology to bacterial Very short patch repair (Vsr) endonucleases.[29] The structure has been solved in 2011, confirming the Vsr homology.[30] Is considered part of the PD-(D/E)xk superfamily.[28]
Domain architecture
edit| Hom_end-associated Hint | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| crystal structure of pi-scei miniprecursor | |||||||||
| Identifiers | |||||||||
| Symbol | Hom_end_hint | ||||||||
| Pfam | PF05203 | ||||||||
| Pfam clan | CL0363 | ||||||||
| InterPro | IPR007868 | ||||||||
| SCOP2 | 1gpp / SCOPe / SUPFAM | ||||||||
| |||||||||
| Intein motif of the larger LAGLIDADG Hom_end domain. | |||||||||
The yeast homing endonuclease PI-Sce is a LAGLIDADG-type endonuclease encoded as an intein that splices itself out of another protein (P17255). The high-resolution structure reveals two domains: an endonucleolytic centre resembling the C-terminal domain of Hedgehog proteins, and a Hint domain (Hedgehog/Intein) containing the protein-splicing active site.[31]
See also
edit- REBASE, a comprehensive restriction enzyme database from New England Biolabs with links to related literature.
- List of homing endonuclease cutting sites
- I-CreI homing endonuclease
- Meganucleases
- Restriction enzyme
- Introns and inteins
- Intragenomic conflict: Homing endonuclease genes
- Transposon
References
edit- ^ Edgell DR (February 2009). "Selfish DNA: homing endonucleases find a home". Curr Biol. 19 (3): R115–R117. Bibcode:2009CBio...19.R115E. doi:10.1016/j.cub.2008.12.019. PMID 19211047. S2CID 2380439.
- ^ Jasin M (Jun 1996). "Genetic manipulation of genomonth with rare-cutting endonucleases". Trends Genet. 12 (6): 224–8. doi:10.1016/0168-9525(96)10019-6. PMID 8928227.
- ^ a b Burt A, Koufopanou V (December 2004). "Homing endonuclease genes: the rise and fall and rise again of a selfish element". Curr Opin Genet Dev. 14 (6): 609–15. doi:10.1016/j.gde.2004.09.010. PMID 15531154.
- ^ a b c Belfort M, Roberts RJ (September 1995). "Homing endonucleases: keeping the house in order". Nucleic Acids Res. 25 (17): 3379–88. doi:10.1093/nar/25.17.3379. PMC 146926. PMID 9254693.
- ^ a b Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL (October 2002). "Design, activity, and structure of a highly specific artificial endonuclease". Mol. Cell. 10 (4): 895–905. doi:10.1016/S1097-2765(02)00690-1. PMID 12419232.
- ^ Hirata R, Ohsumk Y, Nakano A, Kawasaki H, Suzuki K, Anraku Y (April 1990). "Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae". J Biol Chem. 265 (12): 6726–33. doi:10.1016/S0021-9258(19)39210-5. PMID 2139027.
- ^ Kane PM, Yamashiro CT, Wolczyk DF, Neff N, Goebl M, Stevens TH (November 1990). "Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H(+)-adenosine triphosphatase". Science. 250 (4981): 651–7. Bibcode:1990Sci...250..651K. doi:10.1126/science.2146742. PMID 2146742.
- ^ Perler FB, Comb DG, Jack WE, Moran LS, Qiang B, Kucera RB, Benner J, Slatko BE, Nwankwo DO, Hempstead SK, Carlow CK, Jannasch H (June 1992). "Intervening sequences in an Archaea DNA polymerase gene". PNAS. 89 (12): 5577–81. Bibcode:1992PNAS...89.5577P. doi:10.1073/pnas.89.12.5577. PMC 49335. PMID 1608969.
- ^ a b c Jurica MS, Monnat RJ, Stoddard BL (October 1998). "DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI" (PDF). Mol. Cell. 2 (4): 469–76. doi:10.1016/S1097-2765(00)80146-X. PMID 9809068.
- ^ Gimble FS, Wang J (October 1996). "Substrate recognition and induced DNA distortion by the PI-SceI endonuclease, an enzyme generated by protein splicing". J Mol Biol. 263 (2): 163–80. doi:10.1006/jmbi.1996.0567. PMID 8913299.
- ^ Argast GM, Stephens KM, Emond MJ, Monnat RJ (July 1998). "I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment". J Mol Biol. 280 (3): 345–53. doi:10.1006/jmbi.1998.1886. PMID 9665841.
- ^ Shibata T, Nakagawa K, Morishima N (1995). "Multi-site-specific endonucleases and the initiation of homologous genetic recombination in yeast". Adv Biophys. 31: 77–91. doi:10.1016/0065-227X(95)99384-2. PMID 7625280.
- ^ Zimmerly S, Guo H, Eskes R, Yang J, Perlman PS, Lambowitz AM (November 1995). "A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility". Cell. 83 (4): 529–38. doi:10.1016/0092-8674(95)90092-6. PMID 7585955. S2CID 10456475.
- ^ Linn, Stuart M; Lloyd, R Stephen; Roberts, Richard J (December 1993). Nucleases. Cold Spring Harbor Press. pp. 35–88. ISBN 978-0-87969-426-5.
- ^ a b Linn, Stuart M; Lloyd, R Stephen; Roberts, Richard J (December 1993). Nucleases. Cold Spring Harbor Press. pp. 89–109. ISBN 978-0-87969-426-5.
- ^ Roberts RJ, Macelis D (January 1997). "REBASE-restriction enzymes and methylases". Nucleic Acids Res. 25 (1): 248–62. doi:10.1093/nar/25.1.248. PMC 146408. PMID 9016548.
- ^ Lambowitz AM, Belfort M (1993). "Introns as mobile genetic elements". Annu Rev Biochem. 62: 587–622. doi:10.1146/annurev.bi.62.070193.003103. PMID 8352597.
- ^ Linn, Stuart M; Lloyd, R Stephen; Roberts, Richard J (December 1993). Nucleases. Cold Spring Harbor Press. pp. 111–143. ISBN 978-0-87969-426-5.
- ^ Wilson GG (December 1988). "Cloned restriction-modification systems—a review". Gene. 74 (1): 281–9. doi:10.1016/0378-1119(88)90304-6. PMID 3074014.
- ^ Heath, P.; et al. (June 1997). "The structure of I-Crel, a group I intron-encoded homing endonuclease". Nature Structural Biology. 4 (6): 468–476. doi:10.1038/nsb0697-468. PMID 9187655. S2CID 12261983.
- ^ Duan, X. (May 1997). "Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity". Cell. 89 (4): 555–564. doi:10.1016/S0092-8674(00)80237-8. PMID 9160747. S2CID 14156646.
- ^ Van Roey, P.; Fox, KM; et al. (July 2001). "Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate". EMBO J. 20 (14): 3631–3637. doi:10.1093/emboj/20.14.3631. PMC 125541. PMID 11447104.
- ^ Van Roey, P.; Kowalski, Joseph C.; et al. (July 2002). "Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI". Nature Structural Biology. 9 (11): 806–811. doi:10.1038/nsb853. PMID 12379841. S2CID 24856337.
- ^ Flick, K.; et al. (July 1998). "DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI". Nature. 394 (6688): 96–101. Bibcode:1998Natur.394...96F. doi:10.1038/27952. PMID 9665136. S2CID 4427957.
- ^ Hafez, M; Hausner, G (August 2012). "Homing endonucleases: DNA scissors on a mission". Genome. 55 (8): 553–69. doi:10.1139/g2012-049. PMID 22891613. S2CID 29183470.
- ^ Shen, B.W.; et al. (September 2004). "DNA binding and cleavage by the HNH homing endonuclease I-HmuI". J. Mol. Biol. 342 (1): 43–56. doi:10.1016/j.jmb.2004.07.032. PMID 15313606. S2CID 15990707.
- ^ Zhao, L.; et al. (May 2007). "The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif". EMBO Journal. 26 (9): 2432–2442. doi:10.1038/sj.emboj.7601672. PMC 1864971. PMID 17410205.
- ^ a b Steczkiewicz, K; Muszewska, A; Knizewski, L; Rychlewski, L; Ginalski, K (August 2012). "Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily". Nucleic Acids Research. 40 (15): 7016–45. doi:10.1093/nar/gks382. PMC 3424549. PMID 22638584.
- ^ Dassa, B.; et al. (March 2009). "Fractured genes: a novel genomic arrangement involving new split inteins and a new homing endonuclease family". Nucleic Acids Research. 37 (8): 2560–2573. doi:10.1093/nar/gkp095. PMC 2677866. PMID 19264795.
- ^ Taylor, GK; Heiter, DF; Pietrokovski, S; Stoddard, BL (December 2011). "Activity, specificity and structure of I-Bth0305I: a representative of a new homing endonuclease family". Nucleic Acids Research. 39 (22): 9705–19. doi:10.1093/nar/gkr669. PMC 3239194. PMID 21890897.
- ^ Moure CM, Gimble FS, Quiocho FA (October 2002). "Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence". Nat. Struct. Biol. 9 (10): 764–70. doi:10.1038/nsb840. PMID 12219083. S2CID 40192379.
External links
edit- Perler FB. "InBase". Archived from the original on 2010-08-02. Retrieved 2010-08-09.
The Intein Database and Registry (from New England Biolabs)
- Perler FB (January 2002). "InBase: the Intein Database". Nucleic Acids Res. 30 (1): 383–4. doi:10.1093/nar/30.1.383. PMC 99080. PMID 11752343.