Cyanophages are viruses that infect cyanobacteria, also known as Cyanophyta or blue-green algae. Cyanobacteria are a phylum of bacteria that obtain their energy through the process of photosynthesis.[1][2] Although cyanobacteria metabolize photoautotrophically like eukaryotic plants, they have prokaryotic cell structure. Cyanophages can be found in both freshwater and marine environments.[3] Marine and freshwater cyanophages have icosahedral heads, which contain double-stranded DNA, attached to a tail by connector proteins.[4] The size of the head and tail vary among species of cyanophages. Cyanophages infect a wide range of cyanobacteria and are key regulators of the cyanobacterial populations in aquatic environments, and may aid in the prevention of cyanobacterial blooms in freshwater and marine ecosystems. These blooms can pose a danger to humans and other animals, particularly in eutrophic freshwater lakes. Infection by these viruses is highly prevalent in cells belonging to Synechococcus spp. in marine environments, where up to 5% of cells belonging to marine cyanobacterial cells have been reported to contain mature phage particles.[5]
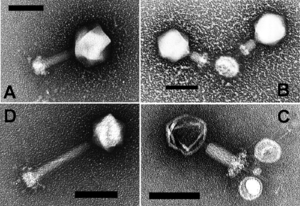
The first described cyanophage LPP-1, was reported by Safferman and Morris in 1963.[6] Cyanophages are classified within the bacteriophage families Myoviridae (e.g. AS-1, N-1), Podoviridae (e.g. LPP-1) and Siphoviridae (e.g. S-1).[6]
Nomenclature
editThe following three families of cyanophages have been recognized by the International Committee on Taxonomy of Viruses (ICTV): Myoviridae, Siphoviridae and Podoviridae; all contain double-stranded DNA.[7] Initially, cyanophages were named after their hosts. However, the ability of cyanophages to infect multiple hosts and lack of a universal naming system can cause difficulties with their taxonomic classification.[8] Many other classification systems used serological, morphological, or physiological properties.[9][10] Currently, the suggested procedure of naming strains is as follows: Cyanophage Xx-YYZaa, where Xx is the first two letters of the genus and species names of the host that the type specimen phage is found in, YY is the origin of the specimen, Z is the virus family, and aa is the reference number of the virus.[3]
Morphology
editLike all other tailed bacteriophages cyanophages have a tail and a protein capsid surrounding genetic material. The double-stranded DNA is approximately 45 kilo-base-pairs long and in some cyanophages encodes photosynthetic genes, an integrase, or genes involved with phosphate metabolism (phosphate-inducible).[11] The tail binds the virus to the host cell and transfers viral DNA to the host cell upon infection. Based on morphological characteristics, cyanophages are placed into the families Myoviridae, Podoviridaeand Siphoviridae, and although not formally recognized by the International Committee on Taxonomy of Viruses, historically cyanophages have been further classified into as a Cyanomyovirus, Cyanopodovirus or Cyanostylovirus based on which of the three families in which they are grouped.[8]
Cyanomyovirus
editThe type species for Cyanomyovirus of the family Myoviridae is Cyanophage AS-1, which was isolated from a waste stabilization pond,[12] and was also the first genus recognized.[13] The tails have been observed as either contractile or noncontractile with lengths of 20 to 244 nm, widths of 15 to 23 nm, and a shrinking range of 93 nm.[14][3] Cyanophages generally have isometric hexagonal heads with diameters ranging from 55 to 90 nm.[14][3] There is large morphological variation in this group, which suggests that they infect a variety of host species.[15] At the point of attachment between the long tail and the head there is a base plate where short pins are attached, a contractile sheath, and an internal core, similar to other bacteriophages in the Myoviridae.[12]
Cyanopodovirus
editCyanopodovirus, within the Podoviridae, are present in both fresh and marine water.[16] The type specimen of cyanopodovirus is Cyanophage LPP-1, which infects Lyngbya, Plectonema and Phormidium.[17] Their capsids are polyhedrons that appear hexagonal in 2-D.[14] The tails are hollow with sixfold radial symmetry made of rings of six subunits with unknown orientation.[14] Similar to cyanomyoviruses, they can be found in waste-stabilization ponds and have isometric capsids of similar size but shorter tails.[3]
Cyanostylovirus
editCyanostylovirus belong to the family Siphoviridae, where the type species is Cyanophage S-1, which is known to infect Synechococcus.[3] Cyanostyloviridae have smaller (50 nm in diameter) isometric capsids than the previous genera but longer tails (140 nm).[18] Other genera in this family have tails that range from 200 to 300 nm in length.[15]
Host
editThe host range of cyanophages is very complex and is thought to play an important role in controlling cyanobacterial populations.[1] Freshwater cyanophages have been reported to infect hosts in more than one genus although this may also reflect problems in the taxonomic classification of their hosts. Nonetheless, they have been classified into three major groups based on the taxonomy of their host organism.[1][3]
LPP group
editThe first group is LPP, which belongs to the cyanopodoviruses.[1] This group of viruses includes the original cyanophage isolate that infected "blue-green algae".[13][3] Cyanophages in this group are easy to isolate from the environment.[3] They carry short non-contractile tails and cause lysis of several species within three genera of cyanobacteria: Lyngbya, Plectonema and Phormidium.[3] Thus, the name LPP was derived from the three genera of hosts that they infect.[13] LPP-1 and LPP-2 are two major types of LPP cyanhophages.[19] This group of cyanophages has the same host same range; however, their serum and other body fluids are not the same.[19]
AS and SM group
editThe AS and SM groups represent the third group of cyanophages classified based on host range.[1] This group of viruses is said to be the “new blue-green algae” and infects unicellular forms of cyanobacteria.[3][20][12] The myovirus AS-1 infects Anacystis nidulans,[21] Synechococcus cedrorum, Synechococcus elongatus and Microcystis aeruginosa.[3] Similarly, the unicellular blue-green algae Synechococcus elongatus and Microcystis aeruginosa are infected by the podovirus SM-1.[3][22] There is a new SM-group of virus, known as SM-2, which also lyses Microcystis aeruginosa[22].
A, AN, N and NP group
editCyanophages classified in the groups A, AN, N and NP represent a second group of cyanophages classified based on host range.[18][1][23][24] They play an important role in infecting and causing lysis of members of the genera Nostoc, Anabaena and Plectonema.[1] The A-group of the virus causes lysis and infects Anabaena species.[3] Similarly, the host range of the AN group includes both Anabaena and Nostoc species; whereas, the N group of viruses infects Nostoc species only and includes Cyanophage N-1.[3] Cyanophage N-1 is remarkable in that it encodes a functional CRISPR array that may provide immunity to the host to infection by competing cyanophages.[25] Lastly, cyanobacterial isolates of Nostoc and Plectonema species are infected by the NP group of viruses.[3] These cyanobacterial isolates closely relate to the taxonomic group of Nostoc.[3] They all have a broad host range and mutations are noticeable in these groups of viruses.[3]
Replication
editCyanophage replication has two dominant cycles: the lytic cycle and the lysogenic cycle. Viral nucleic-acid replication and immediate synthesis of virus-encoded protein is considered to be the lytic cycle. Phages are considered lytic if they only have the capacity to enter the lytic cycle; whereas, temperate phage can either enter the lytic cycle or become stably integrated with the host genome and enter the lysogenic cycle.[26] To meet the metabolic demand of replication, viruses recruit a multitude of strategies to sequester nutrients from their host. One such technique is to starve their host cell. This is done by inhibiting the host cells CO2 fixation, which enables the cyanophage to recruit photosynthetically formed redox and ATP from the host cell to meet their nucleotide and metabolic response.[27] Many cyanophages contain genes known as viral-encoded auxiliary metabolic genes (AMGs), which encode critical, rate-limiting steps of the host organism.[27] AMGs encode genes for the pentose phosphate pathway, phosphate acquisition, sulfur metabolism, and DNA/RNA processing; these genes interfere with the metabolism of the host cell. Metagenomic analysis highly supports the notion that these genes promote viral replication through the degradation of host DNA and RNA, as well as a shift in host-cell metabolism to nucleotide biosynthesis.[27] Cyanophages also use these genes to maintain host photosynthesis through the progression of the infection, shuttling the energy away from carbon fixation to anabolism, which the virus takes advantage of.[28] AMGs also code for proteins, which aid in the repair of the host photosystem, which is susceptible to photodegradation.[28] One such example is the D1 proteins which replace the host cells D1 protein when it becomes damaged.[28] The virus up-regulates photosynthesis, which leads to an increased rate of D1 protein degradation, the host cell alone can not efficiently replace these proteins so the cyanophage replaces them for the host cell, allowing it to continue providing energy for the cyanophage replication cycle.[28]
It is evident that cyanophage replication is heavily dependent on the diel cycle. The first step in the infectious cycle is for the cyanophage to make contact and bind to the cyanobacteria, this adsorption process is heavily dependent on light intensity.[29] Field studies also show that the infection and replication of cyanophages is directly or indirectly synchronized with the light-dark cycle.[29]
Adherence
editCyanophages like other bacteriophages rely on Brownian motion to collide with bacteria, and then use receptor binding proteins to recognize cell surface proteins, which leads to adherence. Viruses with contractile tails then rely on receptors found on their tails to recognize highly conserved proteins on the surface of the host cell.[30] Cyanophages also have several surface proteins with Ig-like domains, which are used for adherence.[30]
Some cyanophages also produce a horn like structure, which projects from the vertex opposite of the tail.[31] The horn like structure is hypothesized to aid in attachment to cells in the natural environment; however, this has not been confirmed.[31]
Lytic cycle
editCyanophages can undergo both the lytic and lysogenic cycles depending on the viruses and their environment.[32][33] In one study on cyanomyoviruses infecting marine Synechococcus sp., the lytic phase was shown to last approximately 17 hours with the average number of viruses produced for each cell that was lysed (burst size) ranging from 328 under high light to 151 under low light.[34] There is evidence supporting the premise that there is a correlation between light intensity and burst size.[29] Studies show that cyanophage replication is driven by energy from photosynthetic metabolism of the host cell.[35] Lysing of the host cell tends to occur after the completion of host DNA replication and immediately prior to cell division.[35] This is due to the increased availability of resources for the replication of viral particles.[citation needed]
Ecological Significance
editEcosystem
editCertain cyanophages infect and burst Prochlorococcus, the world's smallest and most abundant primary producers.[37][11] Marine cyanophages of the family Myoviridae help regulate primary production mainly through infection of Synechococcus spp.[3] The other two families, Podoviridae and Siphoviridae, are usually found in freshwater ecosystems.[3] In coastal oceans, abundance of viruses infecting Synechococcus spp. can reach >106 mL−1 and 105 g−1 in sediments.[3] An estimated 3% of Synechococcus are removed daily by cyanophages.[3] Cyanophages are widely distributed both throughout the water column and geographically.[3][37][38] Cyanophage populations have been found to inhabit microbial mats in the Arctic through metagenomic analysis and hypersaline lagoons.[38][4] They can withstand temperatures ranging from 12 to 30 °C and salinities of 18-70 ppt.[4] The DNA of cyanophages is susceptible to UV degradation but can be restored in host cells through a process called "photoreactivation".[39] The viruses cannot move independently and must rely on currents, mixing, and host cells to transport them. Viruses cannot actively target their hosts and must wait to encounter them. The higher probability of collision may explain why cyanophages of the Myoviridae family primarily infect one of the most abundant cyanobacteria, Synechoccocus.[3] Evidence of seasonal co-variation between the phages and hosts, in addition to an increase in cyanophages above a threshold of 103 to 104 Synechococcus mL−1, may suggest a “kill-the-winner” dynamic.[3]
Biological and physical impact
editMembers of the genus Synechococcus contribute ~25% to photosynthetic primary productivity in the ocean, having significant bottom-up effect on higher trophic levels.[40] The dissolved organic matter (DOM) released from viral lysis by cyanophages can be shunted into the microbial loop where it is recycled or rejected by heterotrophic bacteria to form recalcitrant matter that is eventually buried in sediment.[40][41] This is an important step in atmospheric carbon sequestration, commonly referred to as the biological pump, and maintenance of other biogeochemical cycles.[40]
Cyanobacteria perform oxygenic photosynthesis which is thought to be the origin of atmospheric oxygen approximately 2.5Ga ago.[42] Population, and therefore, rate of oxygen evolution can be regulated by cyanophages. In certain species of cyanobacteria, such as Trichodesmium that perform nitrogen fixation, cyanophages are capable of increasing the supply rate of bioavailable organic nitrogen through lysis.[43][44]
Cyanophages also infect bloom-forming cyanobacteria that can be toxic to health of humans and other animals through the production of microcystin and cause eutrophication, leading to oxygen minimum zones. Cyanophages can infect and kill four common bloom-forming cyanobacteria: Lyngbya birgei, Anabaena circinalis, Anabaena flosaquae, and Microcystis aeruginosa,[26] and thus may be able to prevent harmful algal blooms under normal conditions. Blooms cause problems ecologically, economically, and in freshwater systems, adversely affect the quality of drinking water.[45] Spikes in cyanobacteria populations are usually brought on by nutrient increases due to run-off from fertilizers, dust, and sewage.[46] By killing hosts, cyanophages may help restore ecosystems to their natural balance.[citation needed]
In addition to regulating population size, cyanophages likely influence phylogenetic composition by allowing other phytoplankton normally inhibited by cyanobacteria to grow.[46] The specificity with which cyanophages target various hosts also affects community structure. Due to the lysogenic phase of their replication cycle, cyanophages may behave as mobile genetic elements for genetic diversification of their hosts through horizontal gene transfer.[47][27] Whether the lytic or lysogenic phase dominates in a given area has been hypothesized to depend on eutrophic or oligotrophic conditions, respectively.[41] Increase in number of encounters is directly related to an increase in rate of infection providing more opportunity for selective pressure, making coastal Synechococcus more resistant to viral infection than their off-shore counterparts.[3]
References
edit- ^ a b c d e f g Xia H, Li T, Deng F, Hu Z (2013-10-01). "Freshwater cyanophages". Virologica Sinica. 28 (5): 253–259. doi:10.1007/s12250-013-3370-1. ISSN 1674-0769. PMC 8208336. PMID 24132756.
- ^ Whitton BA, Potts M (2000). The Ecology of Cyanobacteria: Their Diversity in Time and Space. Boston: Kluwer Academic. pp. 563–589. ISBN 978-0-7923-4735-4.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z Suttle CA (2000-01-01). "Cyanophages and Their Role in the Ecology of Cyanobacteria". In Whitton BA, Potts M (eds.). The Ecology of Cyanobacteria. Springer Netherlands. pp. 563–589. doi:10.1007/0-306-46855-7_20. ISBN 978-0-7923-4735-4.
- ^ a b c Suttle CA, Chan AM (1993). "Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, . morphology, cross-infectivity and growth characteristics". Marine Ecology Progress Series. 92: 99–109. Bibcode:1993MEPS...92...99S. doi:10.3354/meps092099.
- ^ Proctor LM, Fuhrman JA (1990). "Viral mortality of marine bacteria and cyanobacteria". Nature. 343 (6253): 60–62. Bibcode:1990Natur.343...60P. doi:10.1038/343060a0. S2CID 4336344.
- ^ a b Sarma TA. 'Cyanophages' in Handbook of Cyanobacteria (CRC Press; 2012) (ISBN 1466559411)
- ^ King A, Lefkowitz E, Adams M, Carstens E (2012). Virus Taxonomy Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier. ISBN 978-0-12-384684-6.
- ^ a b Safferman R, Cannon R, Desjardins P, Gromov B, Haselkorn R, Sherman L, Shilo M (1983). "Classification and Nomenclature of Viruses of Cyanobacteria". Intervirology. 19 (2): 61–66. doi:10.1159/000149339. PMID 6408019.
- ^ Gibbs AJ (2005). Molecular Bases of Virus Evolution. Cambridge University Press. ISBN 978-0-521-02289-7.
- ^ Stanier R (1971). "Purification and properties of unicellular blue-green algae (order Chroocococcales)". Bacteriological Reviews. 35 (2): 171–205. doi:10.1128/MMBR.35.2.171-205.1971. PMC 378380. PMID 4998365.
- ^ a b Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW (2005-04-19). "Three Prochlorococcus Cyanophage Genomes: Signature Features and Ecological Interpretations". PLOS Biology. 3 (5): e144. doi:10.1371/journal.pbio.0030144. ISSN 1545-7885. PMC 1079782. PMID 15828858.
- ^ a b c Safferman R, Diener T, Desjardins P, Morris M (1972). "Isolation and characterization of AS-1, a phycovirus infecting the blue-green algae, Anacystis nidulans and Synechococcus cedrorum". Virology. 47 (1): 105–113. doi:10.1016/0042-6822(72)90243-7. PMID 4110125.
- ^ a b c Safferman RS, Morris ME (1964). "Growth Characteristics of the Blue-Green Algal Virus LPP-1". J. Bacteriol. 88 (3): 771–775. doi:10.1128/JB.88.3.771-775.1964. PMC 277376. PMID 14208517.
- ^ a b c d Padan E, Shilo M (1973). "Cyanophages- viruses attacking blue-green algae". Bacteriological Reviews. 37 (3): 343–370. doi:10.1128/MMBR.37.3.343-370.1973. PMC 413822. PMID 4202147.
- ^ a b Gromov B (1983). "Cyanophages". Annales de l'Institut Pasteur / Microbiologie. 134 (1): 43–59. doi:10.1016/s0769-2609(83)80096-9. PMID 6416127.
- ^ Hu NT, Thiel T, Giddings TH, Wolk C (1981). "New Anabaena and Nostoc cyanophages from sewage settling ponds". Virology. 114 (1): 236–246. doi:10.1016/0042-6822(81)90269-5. PMID 6269286.
- ^ Schneider IR, Diener TO, Safferman RS (1964-05-29). "Blue-Green Algal Virus LPP-1: Purification and Partial Characterization". Science. 144 (3622): 1127–1130. Bibcode:1964Sci...144.1127S. doi:10.1126/science.144.3622.1127. ISSN 0036-8075. PMID 14148431. S2CID 45125402.
- ^ a b Adolph KW, Haselkorn R (1973). "Isolation and characterization of a virus infecting a blue-green alga of the genus Synechococcus". Virology. 54 (1): 230–236. doi:10.1016/0042-6822(73)90132-3. PMID 4197413.
- ^ a b JOHNSON DW, POTTS M (1985). "Host Range of LPP Cyanophages". International Journal of Systematic Bacteriology. 35: 76–78. doi:10.1099/00207713-35-1-76.
- ^ SAFFERMAN R, Schneider I, Steere R, MORRIS M, DIENER T (1969). "Phycovirus SM-1: A virus infecting unicellular blue-green algae". Virology. 37 (3): 386–397. doi:10.1016/0042-6822(69)90222-0. PMID 5777559.
- ^ Orkwiszewski KG, Kaney AR (June 1974). "Genetic transformation of the blue-green bacterium, Anacystis nidulans". Arch Mikrobiol. 98 (1): 31–37. Bibcode:1974ArMic..98...31O. doi:10.1007/BF00425265. PMID 4209657. S2CID 5635245.
- ^ a b Fox JA, Booth S, Martin E (1976). "Cyanophage SM-2: A new blue-green algal virus". Virology. 73 (2): 557–560. doi:10.1016/0042-6822(76)90420-7. PMID 8869.
- ^ MURADOV MM (1990). "COMPARATIVE-STUDY OF NP-1T CYANOPHAGES, WHICH LYSOGENIZE NITROGEN-FIXING BACTERIA OF THE GENERA NOSTOC AND PLECTONEMA". Microbiology. 59 (5): 558–563.
- ^ Kozayakov S (1977). "Cyanophages of the series A(L) specific for the blue-green alga Anabaena variabilis. In". Experimental Algology: 151–171.
- ^ Chénard C, Wirth JF, Suttle CA (2016), "Viruses Infecting a freshwater filamentous cyanobacterium (Nostoc sp.) encode a functional CRISPR array and a proteobacterial DNA polymerase B", mBio, 7 (3): e00667-16, doi:10.1128/mBio.00667-16, PMC 4916379, PMID 27302758
- ^ a b Jassim SA, Limoges RG (2013-10-01). "Impact of external forces on cyanophage–host interactions in aquatic ecosystems". World Journal of Microbiology and Biotechnology. 29 (10): 1751–1762. doi:10.1007/s11274-013-1358-5. ISSN 0959-3993. PMID 23619821. S2CID 24177191.
- ^ a b c d Kaplan A (2016). "Cyanophages: Starving the Host to Recruit Resources". Cell. 26 (12): R511–R513. Bibcode:2016CBio...26.R511K. doi:10.1016/j.cub.2016.04.030. PMID 27326715.
- ^ a b c d Frank JA, Lorimer D, Youle M, Witte P, Craig T, Abendroth J, Rohwer F, Edwards RA, Segall AM (2013-06-01). "Structure and function of a cyanophage-encoded peptide deformylase". The ISME Journal. 7 (6): 1150–1160. Bibcode:2013ISMEJ...7.1150F. doi:10.1038/ismej.2013.4. ISSN 1751-7362. PMC 3660681. PMID 23407310.
- ^ a b c Ni T, Zeng Q (2016-01-01). "Diel Infection of Cyanobacteria by Cyanophages". Frontiers in Marine Science. 2. doi:10.3389/fmars.2015.00123.
- ^ a b Fokine A, Rossmann MG (2014-01-01). "Molecular architecture of tailed double-stranded DNA phages". Bacteriophage. 4 (1): e28281. doi:10.4161/bact.28281. PMC 3940491. PMID 24616838.
- ^ a b Raytcheva DA, Haase-Pettingell C, Piret J, King JA (2014-02-15). "Two Novel Proteins of Cyanophage Syn5 Compose Its Unusual Horn Structure". Journal of Virology. 88 (4): 2047–2055. doi:10.1128/JVI.02479-13. ISSN 0022-538X. PMC 3911526. PMID 24307583.
- ^ McDaniel L, Houchin LA, Williamson SJ, Paul JP (2002). "Plankton blooms - Lysogeny in marine Synechococcus". Nature. 415 (6871): 496. Bibcode:2002Natur.415..496M. doi:10.1038/415496a. PMID 11823851. S2CID 4418714.
- ^ Ortmann AC, Lawrence JE, Suttle CA (2002). "Lysogeny and lytic viral production during a bloom of the cyanobacterium Synechococcus spp". Microbial Ecology. 43 (2): 225–231. Bibcode:2002MicEc..43..225O. doi:10.1007/s00248-001-1058-9. PMID 12023729. S2CID 27385452.
- ^ Brigden S (2003). Dynamics of Cyanophage Replication (MSc, Botany). University of British Columbia. doi:10.14288/1.0091069.
- ^ a b Ni T, Zeng Q (2016-01-01). "Diel Infection of Cyanobacteria by Cyanophages" (PDF). Frontiers in Marine Science. 2. doi:10.3389/fmars.2015.00123. ISSN 2296-7745.
- ^ Ian J. Campbell, Jose Luis Olmos Jr., Weijun Xu, Dimithree Kahanda, Joshua T. Atkinson, Othneil Noble Sparks, Mitchell D. Miller, George N. Phillips Jr., George N. Bennett, Jonathan J. Silberg: Prochlorococcus phage ferredoxin: Structural characterization and electron transfer to cyanobacterial sulfite reductases – Phage Fd characterization and host SIR interactions, in: J. Biol. Chem., ASBMB Publications, 19 May 2020, doi:10.1074/jbc.RA120.013501, PDF. Along with:
- Ocean virus hijacks carbon-storing bacteria, Source: Rice University
- National Science Foundation (NSF), 29 May 2020
- EurekAlert, 26 May 2020
- Ocean ScienceDaily, 26 May 2020
- Beneath the Ocean’s Surface, a Virus Is Hijacking the Most Abundant Organism on Earth, on: SciTechDaily, 6 June 2020. Source: Rice University
- ^ a b Partensky F, Hess WR, Vaulot D (1999-03-01). "Prochlorococcus, a Marine Photosynthetic Prokaryote of Global Significance". Microbiology and Molecular Biology Reviews. 63 (1): 106–127. doi:10.1128/MMBR.63.1.106-127.1999. ISSN 1092-2172. PMC 98958. PMID 10066832.
- ^ a b Varin T, Lovejoy C, Jungblut AD, Vincent WF, Corbeila J (2010). "Metagenomic profiling of Arctic microbial mat communities as nutrient scavenging and recycling systems". Limnology and Oceanography. 55 (5): 1901–1911. Bibcode:2010LimOc..55.1901V. doi:10.4319/lo.2010.55.5.1901. S2CID 55550366.
- ^ Cheng K, Zhao Y, Du X, Zhang Y, Lan S, Shi Z (2007-06-20). "Solar radiation-driven decay of cyanophage infectivity, and photoreactivation of the cyanophage by host cyanobacteria". Aquatic Microbial Ecology. 48 (1): 13–18. doi:10.3354/ame048013.
- ^ a b c Wang K, Wommack KE, Chen F (2011-11-01). "Abundance and Distribution of Synechococcus spp. and Cyanophages in the Chesapeake Bay". Applied and Environmental Microbiology. 77 (21): 7459–7468. Bibcode:2011ApEnM..77.7459W. doi:10.1128/AEM.00267-11. ISSN 0099-2240. PMC 3209163. PMID 21821760.
- ^ a b Weinbauer M (2011). "Virus-Mediated Redistribution and Partitioning of Carbon in the Global Oceans". ResearchGate: 54–56.
- ^ Schirrmeister BE, Antonelli A, Bagheri HC (2011-01-01). "The origin of multicellularity in cyanobacteria". BMC Evolutionary Biology. 11 (1): 45. Bibcode:2011BMCEE..11...45S. doi:10.1186/1471-2148-11-45. ISSN 1471-2148. PMC 3271361. PMID 21320320.
- ^ Bergman B, Sandh G, Lin S, Larsson J, Carpenter EJ (2013-05-01). "Trichodesmium– a widespread marine cyanobacterium with unusual nitrogen fixation properties". FEMS Microbiology Reviews. 37 (3): 286–302. doi:10.1111/j.1574-6976.2012.00352.x. ISSN 0168-6445. PMC 3655545. PMID 22928644.
- ^ Kashyap AK, Rai AN, Singh S (1988-06-01). "Effect of cyanophage N-1 development on nitrogen metabolism of cyanobacterium Nostoc muscorum". FEMS Microbiology Letters. 51 (2–3): 145–148. doi:10.1111/j.1574-6968.1988.tb02986.x. ISSN 0378-1097.
- ^ Beversdorf LJ, Miller TR, McMahon KD (2013-02-06). "The Role of Nitrogen Fixation in Cyanobacterial Bloom Toxicity in a Temperate, Eutrophic Lake". PLOS ONE. 8 (2): e56103. Bibcode:2013PLoSO...856103B. doi:10.1371/journal.pone.0056103. ISSN 1932-6203. PMC 3566065. PMID 23405255.
- ^ a b Fuhrman JA, Suttle CA (1993). "Viruses in marine planktonic systems". Oceanography. 6 (2): 51–63. doi:10.5670/oceanog.1993.14.
- ^ Frost LS, Leplae R, Summers AO, Toussaint A (2005). "Mobile genetic elements: the agents of open source evolution". Nature Reviews Microbiology. 3 (9): 722–732. doi:10.1038/nrmicro1235. PMID 16138100. S2CID 398029.
Further reading
edit- Clokie MR, Mann NH (Dec 2006). "Marine cyanophages and light". Environ. Microbiol. 8 (12): 2074–82. Bibcode:2006EnvMi...8.2074C. doi:10.1111/j.1462-2920.2006.01171.x. PMID 17107549.
- Mann NH (Apr 2003). "Phages of the marine cyanobacterial picophytoplankton". FEMS Microbiol. Rev. 27 (1): 17–34. doi:10.1016/S0168-6445(03)00016-0. PMID 12697340.
- Paul JH, Sullivan MB (Jun 2005). "Marine phage genomics: what have we learned?". Current Opinion in Biotechnology. 16 (3): 299–307. doi:10.1016/j.copbio.2005.03.007. PMID 15961031.
- Suttle CA (2000). "Chapter 20: Cyanophages and their role in the ecology of cyanobacteria". In Whitton BA, Potts M (eds.). The Ecology of Cyanobacteria: Their Diversity in Time and Space. Kluwer Academic Publishers. pp. 563–589. ISBN 978-0-7923-4755-2.