Chemotaxis assays are experimental tools for evaluation of chemotactic ability of prokaryotic or eukaryotic cells. A wide variety of techniques have been developed. Some techniques are qualitative - allowing an investigator to approximately determine a cell's chemotactic affinity for an analyte - while others are quantitative, allowing a precise measurement of this affinity.
Quality control
editIn general, the most important requisite is to calibrate the incubation time of the assay both to the model cell and the ligand to be evaluated. Too short incubation time results in no cells in the sample, while too long time perturbs the concentration gradients and measures more chemokinetic than chemotactic responses.
The most commonly used techniques are grouped into two main groups:
Agar-plate techniques
editThis way of evaluation deals with agar-agar or gelatine containing semi-solid layers made prior to the experiment. Small wells are cut into the layer and filled with cells and the test substance. Cells can migrate towards the chemical gradient in the semi solid layer or under the layer as well. Some variations of the technique deal also with wells and parallel channels connected by a cut at the start of the experiment (PP-technique). Radial arrangement of PP-technique (3 or more channels) provides the possibility to compare chemotactic activity of different cell populations or study preference between ligands.[1]
Counting of cells: positive responder cells could be counted from the front of migrating cells, after staining or in native conditions in light microscope.
Two-chamber techniques
editBoyden chamber
editChambers isolated by filters are proper tools for accurate determination of chemotactic behavior. The pioneer type of these chambers was constructed by Boyden.[2] The motile cells are placed into the upper chamber, while fluid containing the test substance is filled into the lower one. The size of the motile cells to be investigated determines the pore size of the filter; it is essential to choose a diameter which allows active transmigration. For modelling in vivo conditions, several protocols prefer coverage of filter with molecules of extracellular matrix (collagen, elastin etc.) Efficiency of the measurements was increased by development of multiwell chambers (e.g. NeuroProbe), where 24, 96, 384 samples are evaluated in parallel. Advantage of this variant is that several parallels are assayed in identical conditions.
Bridge chambers
editIn another setting, the chambers are connected side by side horizontally (Zigmond chamber)[3] or as concentric rings on a slide (Dunn chamber)[4] Concentration gradient develops on a narrow connecting bridge between the chambers and the number of migrating cells is also counted on the surface of the bridge by light microscope. In some cases the bridge between the two chambers is filled with agar and cells have to "glide" in this semisolid layer.
Capillary techniques
editSome capillary techniques provide also a chamber like arrangement, however, there is no filter between the cells and the test substance.[5] Quantitative results are gained by the multiwell type of this probe using 4-8-12-channel pipettes. Accuracy of the pipette and increased number of the parallel running samples is the great advantage of this test.[6]
Counting of cells: positive responder cells are count from the lower chamber (long incubation time) or from the filter (short incubation time). For detection of cells general staining techniques (e.g. trypan blue) or special probes (e.g. mt-dehydrogenase detection with MTT assay) are used. Labelled (e.g. fluorochromes) cells are also used, in some assays cells get labelled during transmigration the filter.
Other techniques
editBesides the above-mentioned two most commonly used family of techniques, a wide range of protocols were developed to measure chemotactic activity. Some of them are only qualitative, like aggregation tests, where small pieces of agar or filters are placed onto a slide and accumulation of cells around is measured.
In another semiquantitative technique, cells are overlaid the test substance and changes in opalescence of the originally cell-free compartment is recorded during the incubation time.[7]
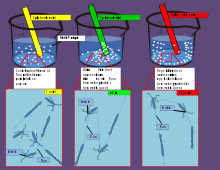
The third frequently used qualitative technique is the T-maze and its adaptations for microplates. In the original version, a container drilled in a peg is filled with cells. Then the peg is twisted and the cells get contact with two other containers filled with different substances. The incubation is stopped by resetting the peg and the cell number is counted from the containers.[8]
Also, lately,[when?] microfluidic devices have been used more and more frequently to test quantitatively, and precisely, for chemotaxis.[9][10][11]
References
edit- ^ Kőhidai L. (1995). "Method for determination of chemoattraction in Tetrahymena pyriformis". Curr Microbiol. 30 (4): 251–3. doi:10.1007/BF00293642. PMID 7765899. S2CID 28989380.
- ^ Boyden, S.V. (1962). "The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes". J Exp Med. 115 (3): 453–66. doi:10.1084/jem.115.3.453. PMC 2137509. PMID 13872176.
- ^ Zigmond S.H. (1977). "Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors". Journal of Cell Biology. 75 (2): 606–616. CiteSeerX 10.1.1.336.4181. doi:10.1083/jcb.75.2.606. PMC 2109936. PMID 264125.
- ^ Zicha D.; Dunn G.A.; Brown A.F. (1991). "A new direct-viewing chemotaxis chamber". J Cell Sci. 99 (4): 769–75. doi:10.1242/jcs.99.4.769. PMID 1770004.
- ^ Leick V.; Helle J. (1983). "A quantitative assay for ciliate chemotaxis". Anal Biochem. 135 (2): 466–9. doi:10.1016/0003-2697(83)90713-3. PMID 6660520.
- ^ Kőhidai, L., Lemberkovits, É. and Csaba, G. (1995). "Molecule dependent chemotactic responses of Tetrahymena pyriformis elicited by volatile oils". Acta Protozool. 34: 181–5.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Koppelhus U.; Hellung-Larsen P.; Leick V. (1994). "An improved quantitative assay for chemokinesis in Tetrahymena". Biol Bull. 187 (1): 8–15. doi:10.2307/1542160. JSTOR 1542160. PMID 7918798.
- ^ Van Houten J.; Martel E.; Kasch T. (1982). "Kinetic analysis of chemokinesis of Paramecium". J Protozool. 29 (2): 226–30. doi:10.1111/j.1550-7408.1982.tb04016.x. PMID 7097615.
- ^ Seymour J. R.; J. R. Ahmed; Marcos S. R (2008). "A microfluidic chemotaxis assay to study microbial behavior in diffusing nutrient patches". Limnology and Oceanography: Methods. 6 (9): 477–887. Bibcode:2008LimOM...6..477S. doi:10.4319/lom.2008.6.477. hdl:10453/8517. S2CID 10730867.
- ^ Zhang C.; {\i{}et al.} (2013). "A Sensitive Chemotaxis Assay Using a Novel Microfluidic Device". {\i{}BioMed Research International}. 8: 8211–8.
- ^ Ahmed T.; himizu T. S.; Stocker R. (2010). "Microfluidics for bacterial chemotaxis". {\i{}Integrative Biology}. 2 (11–12): 604–29. doi:10.1039/c0ib00049c. hdl:1721.1/66851. PMID 20967322.